michalisG
commited on
Commit
•
861a9a7
1
Parent(s):
6af449e
adding model
Browse files- Data_preprocessor.joblib +3 -0
- README.md +3 -0
- RandomForestClassifier_Pipeline_explanation.txt +12 -0
- RandomForestClassifier_trained_pipeline.joblib +3 -0
- RandomForestClassifier_trained_pipeline_scores.html +122 -0
- RandomForestClassifier_trained_pipeline_scores.txt +29 -0
- config.json +35 -0
- overfitting_plot.png +0 -0
- pipeline_diagram.html +79 -0
- plot_roc_curve.png +0 -0
- target_labels.pkl +3 -0
- test_plot_classif_report.png +0 -0
Data_preprocessor.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:0f960bd15bd6edf56e0634d10357a2e93f0498aff16d5ac1de94e259123cd202
|
| 3 |
+
size 5716
|
README.md
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
license: mit
|
| 3 |
+
---
|
RandomForestClassifier_Pipeline_explanation.txt
ADDED
|
@@ -0,0 +1,12 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
The Pipeline is using Simple-Imputer to impute the missing values of the data-setbefore pass them to the model.
|
| 2 |
+
|
| 3 |
+
The Pipeline is using One-Hot-Encoding to encode the categorical valuesof the data-set before pass them to model, most of the models need One-hot-encoding, this algorithm transforms the value from a category to numerical.
|
| 4 |
+
|
| 5 |
+
Many machine learning algorithms perform better or converge faster when features are on a relatively similar scale and/or close to normally distributed. This Pipeline uses Standard-Scaler algorithm which follows Standard Normal Distribution (SND). Therefore, it transforms each value in the column to range about the mean 0 and standard deviation 1, ie, each value will be normalised by subtracting the mean and dividing by standard deviation.
|
| 6 |
+
|
| 7 |
+
This Pipeline has a RandomForestClassifier model. This model has been used because the user selected the "Accuracy" option and the machine learning problem is classification.
|
| 8 |
+
|
| 9 |
+
The Grid Search hyper-parameter tuning was used in this Pipeline because the parameter list number was 9 or less, and an exhaustive Grid Search can be run.
|
| 10 |
+
|
| 11 |
+
Columns that have been removed from the training:
|
| 12 |
+
This is the target column: target
|
RandomForestClassifier_trained_pipeline.joblib
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:b9d8698a4b04a78393bd984242a8086765dd371286e5fd08cb1afff7ab0f5372
|
| 3 |
+
size 233235
|
RandomForestClassifier_trained_pipeline_scores.html
ADDED
|
@@ -0,0 +1,122 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
<table border="1" class="dataframe">
|
| 2 |
+
<tbody>
|
| 3 |
+
<tr>
|
| 4 |
+
<th>Problem</th>
|
| 5 |
+
<td>Classification</td>
|
| 6 |
+
</tr>
|
| 7 |
+
<tr>
|
| 8 |
+
<th>Target Column Name</th>
|
| 9 |
+
<td>target</td>
|
| 10 |
+
</tr>
|
| 11 |
+
<tr>
|
| 12 |
+
<th>Model's Name</th>
|
| 13 |
+
<td>RandomForestClassifier</td>
|
| 14 |
+
</tr>
|
| 15 |
+
<tr>
|
| 16 |
+
<th>Accuracy Score</th>
|
| 17 |
+
<td>0.85000</td>
|
| 18 |
+
</tr>
|
| 19 |
+
<tr>
|
| 20 |
+
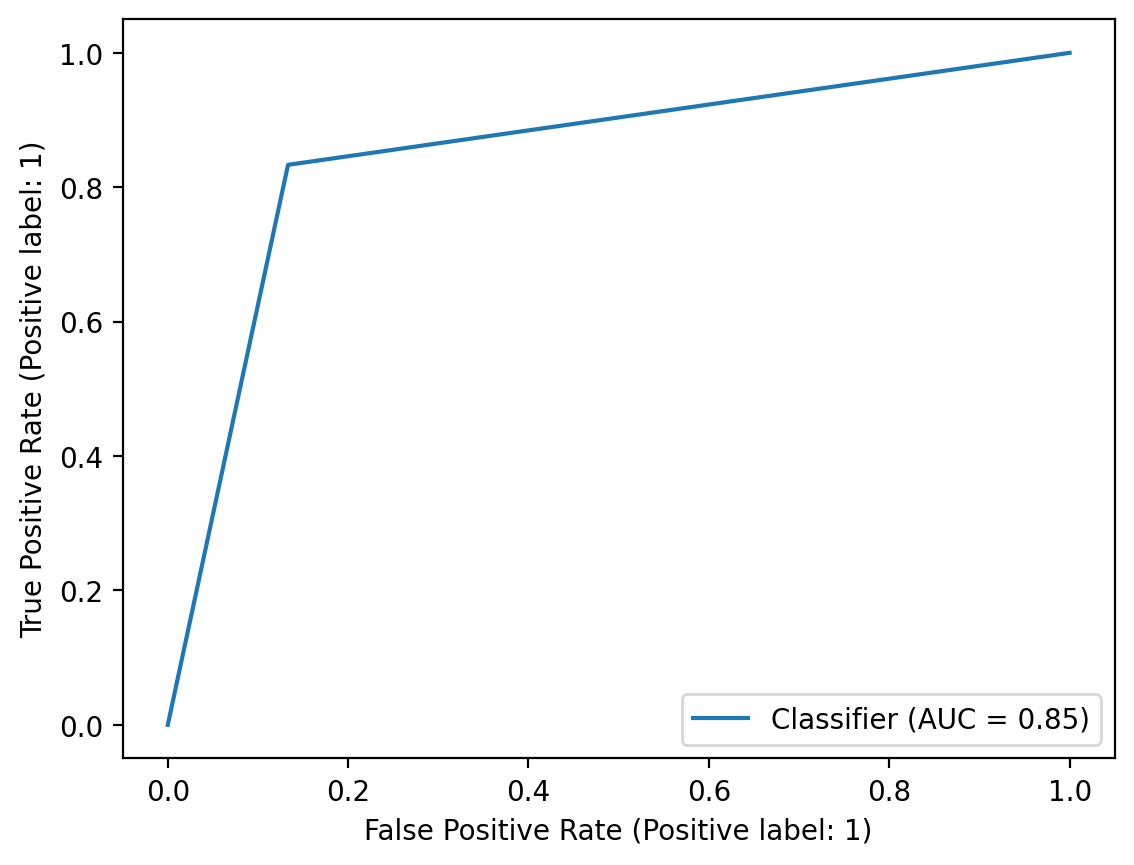
<th>Roc Auc curve</th>
|
| 21 |
+
<td>0.850</td>
|
| 22 |
+
</tr>
|
| 23 |
+
<tr>
|
| 24 |
+
<th>Mean accuracy score of each tested hyperparameter combination</th>
|
| 25 |
+
<td>0.732</td>
|
| 26 |
+
</tr>
|
| 27 |
+
<tr>
|
| 28 |
+
<th>Range of all accuracy scores of each tested hyperparameter combination</th>
|
| 29 |
+
<td>0.708 - 0.792</td>
|
| 30 |
+
</tr>
|
| 31 |
+
<tr>
|
| 32 |
+
<th>Standard Deviation of scores</th>
|
| 33 |
+
<td>0.031</td>
|
| 34 |
+
</tr>
|
| 35 |
+
<tr>
|
| 36 |
+
<th>Standard Deviation < 0.1 * Mean Accuracy scores</th>
|
| 37 |
+
<td>The scores are relatively consistent.</td>
|
| 38 |
+
</tr>
|
| 39 |
+
</tbody>
|
| 40 |
+
</table><font size= 6><p><b> Classification Report:<br><table border="1" class="dataframe">
|
| 41 |
+
<thead>
|
| 42 |
+
<tr style="text-align: right;">
|
| 43 |
+
<th></th>
|
| 44 |
+
<th>precision</th>
|
| 45 |
+
<th>recall</th>
|
| 46 |
+
<th>f1-score</th>
|
| 47 |
+
<th>support</th>
|
| 48 |
+
</tr>
|
| 49 |
+
</thead>
|
| 50 |
+
<tbody>
|
| 51 |
+
<tr>
|
| 52 |
+
<th>N</th>
|
| 53 |
+
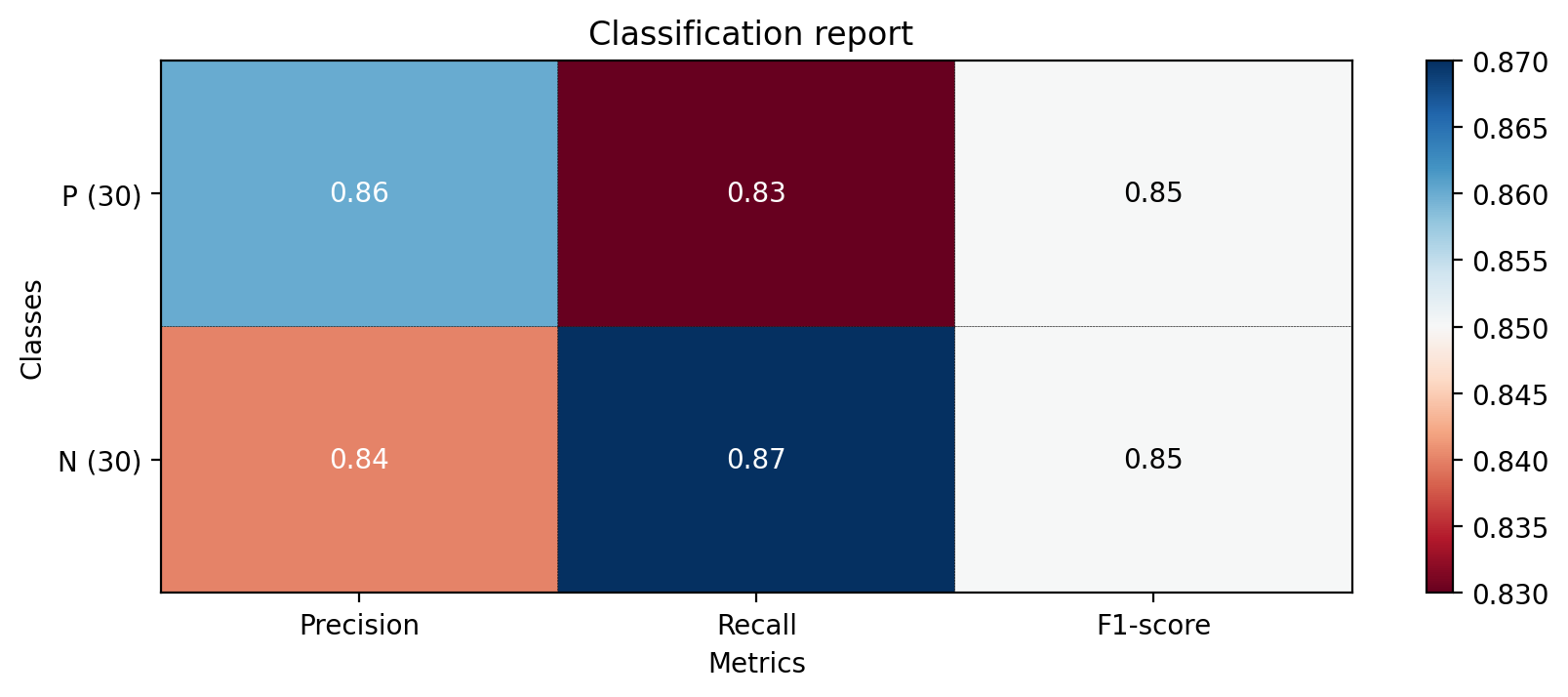
<td>0.838710</td>
|
| 54 |
+
<td>0.866667</td>
|
| 55 |
+
<td>0.852459</td>
|
| 56 |
+
<td>30.00</td>
|
| 57 |
+
</tr>
|
| 58 |
+
<tr>
|
| 59 |
+
<th>P</th>
|
| 60 |
+
<td>0.862069</td>
|
| 61 |
+
<td>0.833333</td>
|
| 62 |
+
<td>0.847458</td>
|
| 63 |
+
<td>30.00</td>
|
| 64 |
+
</tr>
|
| 65 |
+
<tr>
|
| 66 |
+
<th>accuracy</th>
|
| 67 |
+
<td>0.850000</td>
|
| 68 |
+
<td>0.850000</td>
|
| 69 |
+
<td>0.850000</td>
|
| 70 |
+
<td>0.85</td>
|
| 71 |
+
</tr>
|
| 72 |
+
<tr>
|
| 73 |
+
<th>macro avg</th>
|
| 74 |
+
<td>0.850389</td>
|
| 75 |
+
<td>0.850000</td>
|
| 76 |
+
<td>0.849958</td>
|
| 77 |
+
<td>60.00</td>
|
| 78 |
+
</tr>
|
| 79 |
+
<tr>
|
| 80 |
+
<th>weighted avg</th>
|
| 81 |
+
<td>0.850389</td>
|
| 82 |
+
<td>0.850000</td>
|
| 83 |
+
<td>0.849958</td>
|
| 84 |
+
<td>60.00</td>
|
| 85 |
+
</tr>
|
| 86 |
+
</tbody>
|
| 87 |
+
</table><br><img src = "C:\Users\micha\Desktop\Proddis\new_experiment_data\normal_age_version\RandomForestClassifier_Pipeline\test_plot_classif_report.png" alt ="cfg"><br><font size= 6><b> Roc Auc curve figure:</b></font><br><img src = "C:\Users\micha\Desktop\Proddis\new_experiment_data\normal_age_version\RandomForestClassifier_Pipeline\plot_roc_curve.png" alt ="cfg"><br><font size= 6><p><b> Overfit Report:<br><table border="1" class="dataframe">
|
| 88 |
+
<tbody>
|
| 89 |
+
<tr>
|
| 90 |
+
<th>Overfit Report</th>
|
| 91 |
+
<td>The Report is based only on Accuracy</td>
|
| 92 |
+
</tr>
|
| 93 |
+
<tr>
|
| 94 |
+
<th>Train set accuracy score of best pipeline</th>
|
| 95 |
+
<td>0.8661</td>
|
| 96 |
+
</tr>
|
| 97 |
+
<tr>
|
| 98 |
+
<th>Test set accuracy score of best pipeline</th>
|
| 99 |
+
<td>0.8500</td>
|
| 100 |
+
</tr>
|
| 101 |
+
<tr>
|
| 102 |
+
<th>Overfit estimation score of the best pipeline</th>
|
| 103 |
+
<td>0.0161</td>
|
| 104 |
+
</tr>
|
| 105 |
+
<tr>
|
| 106 |
+
<th>Learning Curve scores report</th>
|
| 107 |
+
<td>The Learning Curve is based on Accuracy</td>
|
| 108 |
+
</tr>
|
| 109 |
+
<tr>
|
| 110 |
+
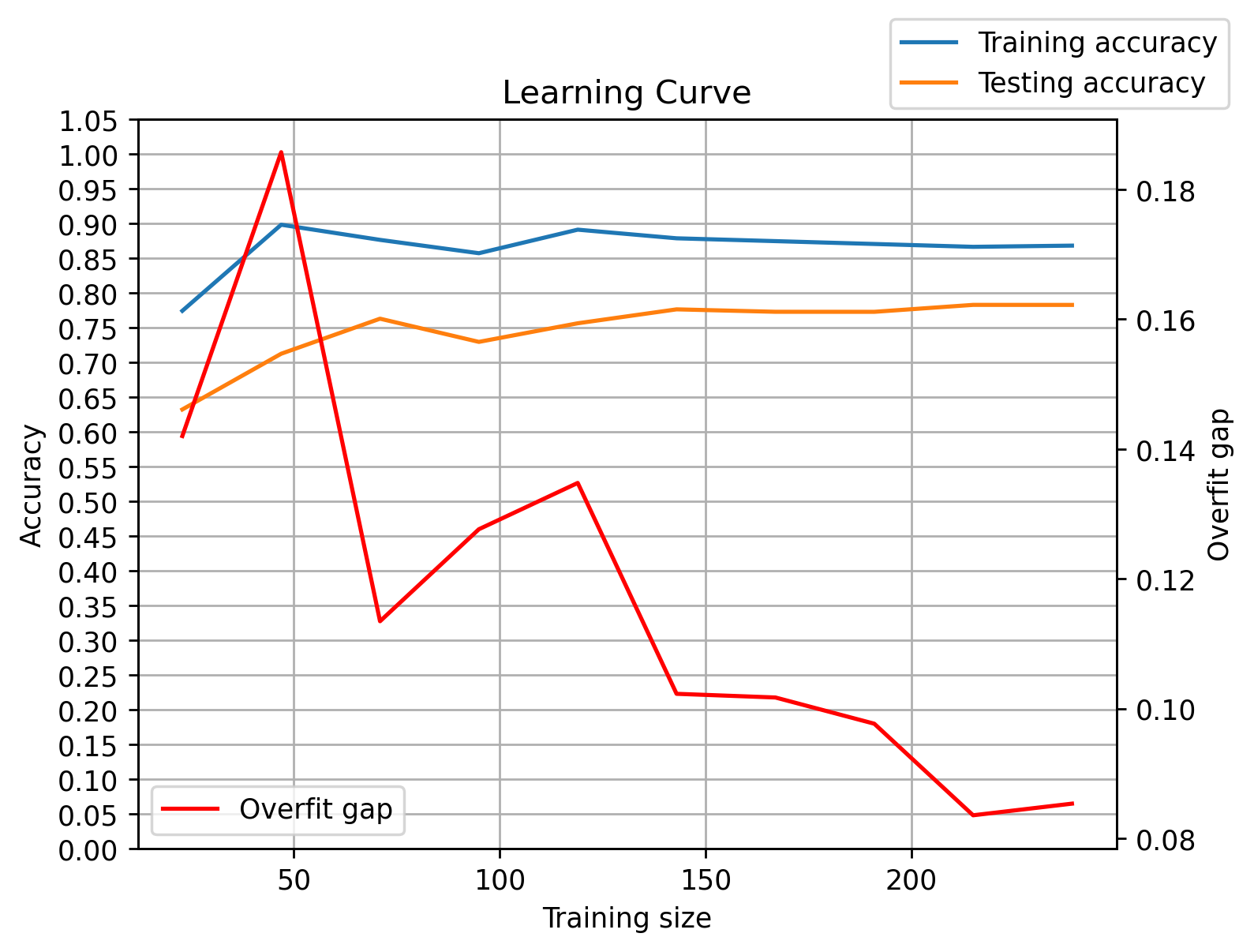
<th>Train set accuracy score of learning curve's last value</th>
|
| 111 |
+
<td>0.87</td>
|
| 112 |
+
</tr>
|
| 113 |
+
<tr>
|
| 114 |
+
<th>Test set accuracy score of learning curve's last value</th>
|
| 115 |
+
<td>0.78</td>
|
| 116 |
+
</tr>
|
| 117 |
+
<tr>
|
| 118 |
+
<th>Overfit gap of learning curve's last value</th>
|
| 119 |
+
<td>0.09</td>
|
| 120 |
+
</tr>
|
| 121 |
+
</tbody>
|
| 122 |
+
</table><br><font size= 6><b> Learning Curve - Overfitting or Underfitting:</b></font><br><img src = "C:\Users\micha\Desktop\Proddis\new_experiment_data\normal_age_version\RandomForestClassifier_Pipeline\overfitting_plot.png" alt ="cfg">
|
RandomForestClassifier_trained_pipeline_scores.txt
ADDED
|
@@ -0,0 +1,29 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Problem: Classification
|
| 2 |
+
model's name: RandomForestClassifier
|
| 3 |
+
precision recall f1-score support
|
| 4 |
+
|
| 5 |
+
N 0.84 0.87 0.85 30
|
| 6 |
+
P 0.86 0.83 0.85 30
|
| 7 |
+
|
| 8 |
+
accuracy 0.85 60
|
| 9 |
+
macro avg 0.85 0.85 0.85 60
|
| 10 |
+
weighted avg 0.85 0.85 0.85 60
|
| 11 |
+
|
| 12 |
+
Accuracy score: 0.85000
|
| 13 |
+
F1 score: 0.847
|
| 14 |
+
Auc score: 0.850
|
| 15 |
+
Overfit estimation with different {:?f}
|
| 16 |
+
0.0
|
| 17 |
+
0.02
|
| 18 |
+
0.016
|
| 19 |
+
0.0161
|
| 20 |
+
Learning curve with different {:?f} for the overall mean value
|
| 21 |
+
0.1
|
| 22 |
+
0.12
|
| 23 |
+
0.117
|
| 24 |
+
0.1174
|
| 25 |
+
Learning curve with different {:?f} for the last value
|
| 26 |
+
0.1
|
| 27 |
+
0.09
|
| 28 |
+
0.085
|
| 29 |
+
0.0854
|
config.json
ADDED
|
@@ -0,0 +1,35 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"model_type": "RandomForestClassifier",
|
| 3 |
+
"expected_features": [
|
| 4 |
+
"age",
|
| 5 |
+
"sex",
|
| 6 |
+
"serum_bilirubin",
|
| 7 |
+
"serum_cholesterol",
|
| 8 |
+
"albumin",
|
| 9 |
+
"alkaline_phosphatase",
|
| 10 |
+
"SGOT",
|
| 11 |
+
"platelets",
|
| 12 |
+
"prothrombin_time"
|
| 13 |
+
],
|
| 14 |
+
"categorical_features": [
|
| 15 |
+
"drug",
|
| 16 |
+
"sex",
|
| 17 |
+
"presence_of_ascites",
|
| 18 |
+
"presence_of_hepatomegaly",
|
| 19 |
+
"presence_of_spiders",
|
| 20 |
+
"presence_of_edema"
|
| 21 |
+
],
|
| 22 |
+
"model_parameters": {
|
| 23 |
+
"criterion": "entropy",
|
| 24 |
+
"max_features": 0.1,
|
| 25 |
+
"min_samples_split": 8,
|
| 26 |
+
"min_samples_leaf": 6,
|
| 27 |
+
"bootstrap": true
|
| 28 |
+
},
|
| 29 |
+
"version": "1.0",
|
| 30 |
+
"preprocessing": {
|
| 31 |
+
"numerical": "median imputation and scaling",
|
| 32 |
+
"categorical": "one-hot encoding",
|
| 33 |
+
"ordinal": "label encoding"
|
| 34 |
+
}
|
| 35 |
+
}
|
overfitting_plot.png
ADDED
|
pipeline_diagram.html
ADDED
|
@@ -0,0 +1,79 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
<style>#sk-container-id-1 {color: black;background-color: white;}#sk-container-id-1 pre{padding: 0;}#sk-container-id-1 div.sk-toggleable {background-color: white;}#sk-container-id-1 label.sk-toggleable__label {cursor: pointer;display: block;width: 100%;margin-bottom: 0;padding: 0.3em;box-sizing: border-box;text-align: center;}#sk-container-id-1 label.sk-toggleable__label-arrow:before {content: "▸";float: left;margin-right: 0.25em;color: #696969;}#sk-container-id-1 label.sk-toggleable__label-arrow:hover:before {color: black;}#sk-container-id-1 div.sk-estimator:hover label.sk-toggleable__label-arrow:before {color: black;}#sk-container-id-1 div.sk-toggleable__content {max-height: 0;max-width: 0;overflow: hidden;text-align: left;background-color: #f0f8ff;}#sk-container-id-1 div.sk-toggleable__content pre {margin: 0.2em;color: black;border-radius: 0.25em;background-color: #f0f8ff;}#sk-container-id-1 input.sk-toggleable__control:checked~div.sk-toggleable__content {max-height: 200px;max-width: 100%;overflow: auto;}#sk-container-id-1 input.sk-toggleable__control:checked~label.sk-toggleable__label-arrow:before {content: "▾";}#sk-container-id-1 div.sk-estimator input.sk-toggleable__control:checked~label.sk-toggleable__label {background-color: #d4ebff;}#sk-container-id-1 div.sk-label input.sk-toggleable__control:checked~label.sk-toggleable__label {background-color: #d4ebff;}#sk-container-id-1 input.sk-hidden--visually {border: 0;clip: rect(1px 1px 1px 1px);clip: rect(1px, 1px, 1px, 1px);height: 1px;margin: -1px;overflow: hidden;padding: 0;position: absolute;width: 1px;}#sk-container-id-1 div.sk-estimator {font-family: monospace;background-color: #f0f8ff;border: 1px dotted black;border-radius: 0.25em;box-sizing: border-box;margin-bottom: 0.5em;}#sk-container-id-1 div.sk-estimator:hover {background-color: #d4ebff;}#sk-container-id-1 div.sk-parallel-item::after {content: "";width: 100%;border-bottom: 1px solid gray;flex-grow: 1;}#sk-container-id-1 div.sk-label:hover label.sk-toggleable__label {background-color: #d4ebff;}#sk-container-id-1 div.sk-serial::before {content: "";position: absolute;border-left: 1px solid gray;box-sizing: border-box;top: 0;bottom: 0;left: 50%;z-index: 0;}#sk-container-id-1 div.sk-serial {display: flex;flex-direction: column;align-items: center;background-color: white;padding-right: 0.2em;padding-left: 0.2em;position: relative;}#sk-container-id-1 div.sk-item {position: relative;z-index: 1;}#sk-container-id-1 div.sk-parallel {display: flex;align-items: stretch;justify-content: center;background-color: white;position: relative;}#sk-container-id-1 div.sk-item::before, #sk-container-id-1 div.sk-parallel-item::before {content: "";position: absolute;border-left: 1px solid gray;box-sizing: border-box;top: 0;bottom: 0;left: 50%;z-index: -1;}#sk-container-id-1 div.sk-parallel-item {display: flex;flex-direction: column;z-index: 1;position: relative;background-color: white;}#sk-container-id-1 div.sk-parallel-item:first-child::after {align-self: flex-end;width: 50%;}#sk-container-id-1 div.sk-parallel-item:last-child::after {align-self: flex-start;width: 50%;}#sk-container-id-1 div.sk-parallel-item:only-child::after {width: 0;}#sk-container-id-1 div.sk-dashed-wrapped {border: 1px dashed gray;margin: 0 0.4em 0.5em 0.4em;box-sizing: border-box;padding-bottom: 0.4em;background-color: white;}#sk-container-id-1 div.sk-label label {font-family: monospace;font-weight: bold;display: inline-block;line-height: 1.2em;}#sk-container-id-1 div.sk-label-container {text-align: center;}#sk-container-id-1 div.sk-container {/* jupyter's `normalize.less` sets `[hidden] { display: none; }` but bootstrap.min.css set `[hidden] { display: none !important; }` so we also need the `!important` here to be able to override the default hidden behavior on the sphinx rendered scikit-learn.org. See: https://github.com/scikit-learn/scikit-learn/issues/21755 */display: inline-block !important;position: relative;}#sk-container-id-1 div.sk-text-repr-fallback {display: none;}</style><div id="sk-container-id-1" class="sk-top-container"><div class="sk-text-repr-fallback"><pre>Pipeline(steps=[('preprocessor',
|
| 2 |
+
ColumnTransformer(transformers=[('num',
|
| 3 |
+
Pipeline(steps=[('SimpleImputer',
|
| 4 |
+
SimpleImputer(strategy='median'))]),
|
| 5 |
+
['age', 'serum_bilirubin',
|
| 6 |
+
'serum_cholesterol',
|
| 7 |
+
'albumin',
|
| 8 |
+
'alkaline_phosphatase',
|
| 9 |
+
'SGOT', 'platelets',
|
| 10 |
+
'prothrombin_time']),
|
| 11 |
+
('cat',
|
| 12 |
+
Pipeline(steps=[('SimpleImputer',
|
| 13 |
+
SimpleImputer(strategy='most_frequent')),
|
| 14 |
+
('OneHotEnc...
|
| 15 |
+
'presence_of_hepatomegaly',
|
| 16 |
+
'presence_of_spiders']),
|
| 17 |
+
('ord',
|
| 18 |
+
Pipeline(steps=[('SimpleImputer',
|
| 19 |
+
SimpleImputer(strategy='most_frequent')),
|
| 20 |
+
('OrdinalEncoder',
|
| 21 |
+
OrdinalEncoder(categories=[['0',
|
| 22 |
+
'0.5',
|
| 23 |
+
'1']]))]),
|
| 24 |
+
['presence_of_edema'])])),
|
| 25 |
+
('RandomForestClassifier',
|
| 26 |
+
RandomForestClassifier(criterion='entropy', max_features=0.1,
|
| 27 |
+
min_samples_leaf=6,
|
| 28 |
+
min_samples_split=8))])</pre><b>In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. <br />On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.</b></div><div class="sk-container" hidden><div class="sk-item sk-dashed-wrapped"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-1" type="checkbox" ><label for="sk-estimator-id-1" class="sk-toggleable__label sk-toggleable__label-arrow">Pipeline</label><div class="sk-toggleable__content"><pre>Pipeline(steps=[('preprocessor',
|
| 29 |
+
ColumnTransformer(transformers=[('num',
|
| 30 |
+
Pipeline(steps=[('SimpleImputer',
|
| 31 |
+
SimpleImputer(strategy='median'))]),
|
| 32 |
+
['age', 'serum_bilirubin',
|
| 33 |
+
'serum_cholesterol',
|
| 34 |
+
'albumin',
|
| 35 |
+
'alkaline_phosphatase',
|
| 36 |
+
'SGOT', 'platelets',
|
| 37 |
+
'prothrombin_time']),
|
| 38 |
+
('cat',
|
| 39 |
+
Pipeline(steps=[('SimpleImputer',
|
| 40 |
+
SimpleImputer(strategy='most_frequent')),
|
| 41 |
+
('OneHotEnc...
|
| 42 |
+
'presence_of_hepatomegaly',
|
| 43 |
+
'presence_of_spiders']),
|
| 44 |
+
('ord',
|
| 45 |
+
Pipeline(steps=[('SimpleImputer',
|
| 46 |
+
SimpleImputer(strategy='most_frequent')),
|
| 47 |
+
('OrdinalEncoder',
|
| 48 |
+
OrdinalEncoder(categories=[['0',
|
| 49 |
+
'0.5',
|
| 50 |
+
'1']]))]),
|
| 51 |
+
['presence_of_edema'])])),
|
| 52 |
+
('RandomForestClassifier',
|
| 53 |
+
RandomForestClassifier(criterion='entropy', max_features=0.1,
|
| 54 |
+
min_samples_leaf=6,
|
| 55 |
+
min_samples_split=8))])</pre></div></div></div><div class="sk-serial"><div class="sk-item sk-dashed-wrapped"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-2" type="checkbox" ><label for="sk-estimator-id-2" class="sk-toggleable__label sk-toggleable__label-arrow">preprocessor: ColumnTransformer</label><div class="sk-toggleable__content"><pre>ColumnTransformer(transformers=[('num',
|
| 56 |
+
Pipeline(steps=[('SimpleImputer',
|
| 57 |
+
SimpleImputer(strategy='median'))]),
|
| 58 |
+
['age', 'serum_bilirubin', 'serum_cholesterol',
|
| 59 |
+
'albumin', 'alkaline_phosphatase', 'SGOT',
|
| 60 |
+
'platelets', 'prothrombin_time']),
|
| 61 |
+
('cat',
|
| 62 |
+
Pipeline(steps=[('SimpleImputer',
|
| 63 |
+
SimpleImputer(strategy='most_frequent')),
|
| 64 |
+
('OneHotEncoder',
|
| 65 |
+
OneHotEncoder(drop='if_binary',
|
| 66 |
+
handle_unknown='ignore',
|
| 67 |
+
sparse=False))]),
|
| 68 |
+
['drug', 'sex', 'presence_of_asictes',
|
| 69 |
+
'presence_of_hepatomegaly',
|
| 70 |
+
'presence_of_spiders']),
|
| 71 |
+
('ord',
|
| 72 |
+
Pipeline(steps=[('SimpleImputer',
|
| 73 |
+
SimpleImputer(strategy='most_frequent')),
|
| 74 |
+
('OrdinalEncoder',
|
| 75 |
+
OrdinalEncoder(categories=[['0',
|
| 76 |
+
'0.5',
|
| 77 |
+
'1']]))]),
|
| 78 |
+
['presence_of_edema'])])</pre></div></div></div><div class="sk-parallel"><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-3" type="checkbox" ><label for="sk-estimator-id-3" class="sk-toggleable__label sk-toggleable__label-arrow">num</label><div class="sk-toggleable__content"><pre>['age', 'serum_bilirubin', 'serum_cholesterol', 'albumin', 'alkaline_phosphatase', 'SGOT', 'platelets', 'prothrombin_time']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-4" type="checkbox" ><label for="sk-estimator-id-4" class="sk-toggleable__label sk-toggleable__label-arrow">SimpleImputer</label><div class="sk-toggleable__content"><pre>SimpleImputer(strategy='median')</pre></div></div></div></div></div></div></div></div><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-5" type="checkbox" ><label for="sk-estimator-id-5" class="sk-toggleable__label sk-toggleable__label-arrow">cat</label><div class="sk-toggleable__content"><pre>['drug', 'sex', 'presence_of_asictes', 'presence_of_hepatomegaly', 'presence_of_spiders']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-6" type="checkbox" ><label for="sk-estimator-id-6" class="sk-toggleable__label sk-toggleable__label-arrow">SimpleImputer</label><div class="sk-toggleable__content"><pre>SimpleImputer(strategy='most_frequent')</pre></div></div></div><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-7" type="checkbox" ><label for="sk-estimator-id-7" class="sk-toggleable__label sk-toggleable__label-arrow">OneHotEncoder</label><div class="sk-toggleable__content"><pre>OneHotEncoder(drop='if_binary', handle_unknown='ignore', sparse=False)</pre></div></div></div></div></div></div></div></div><div class="sk-parallel-item"><div class="sk-item"><div class="sk-label-container"><div class="sk-label sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-8" type="checkbox" ><label for="sk-estimator-id-8" class="sk-toggleable__label sk-toggleable__label-arrow">ord</label><div class="sk-toggleable__content"><pre>['presence_of_edema']</pre></div></div></div><div class="sk-serial"><div class="sk-item"><div class="sk-serial"><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-9" type="checkbox" ><label for="sk-estimator-id-9" class="sk-toggleable__label sk-toggleable__label-arrow">SimpleImputer</label><div class="sk-toggleable__content"><pre>SimpleImputer(strategy='most_frequent')</pre></div></div></div><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-10" type="checkbox" ><label for="sk-estimator-id-10" class="sk-toggleable__label sk-toggleable__label-arrow">OrdinalEncoder</label><div class="sk-toggleable__content"><pre>OrdinalEncoder(categories=[['0', '0.5', '1']])</pre></div></div></div></div></div></div></div></div></div></div><div class="sk-item"><div class="sk-estimator sk-toggleable"><input class="sk-toggleable__control sk-hidden--visually" id="sk-estimator-id-11" type="checkbox" ><label for="sk-estimator-id-11" class="sk-toggleable__label sk-toggleable__label-arrow">RandomForestClassifier</label><div class="sk-toggleable__content"><pre>RandomForestClassifier(criterion='entropy', max_features=0.1,
|
| 79 |
+
min_samples_leaf=6, min_samples_split=8)</pre></div></div></div></div></div></div></div>
|
plot_roc_curve.png
ADDED
|
target_labels.pkl
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:f8195df6a86a0f89a7eb5f6c25b7280b359a00e41526f3891ef5fb482a66049c
|
| 3 |
+
size 24
|
test_plot_classif_report.png
ADDED
|