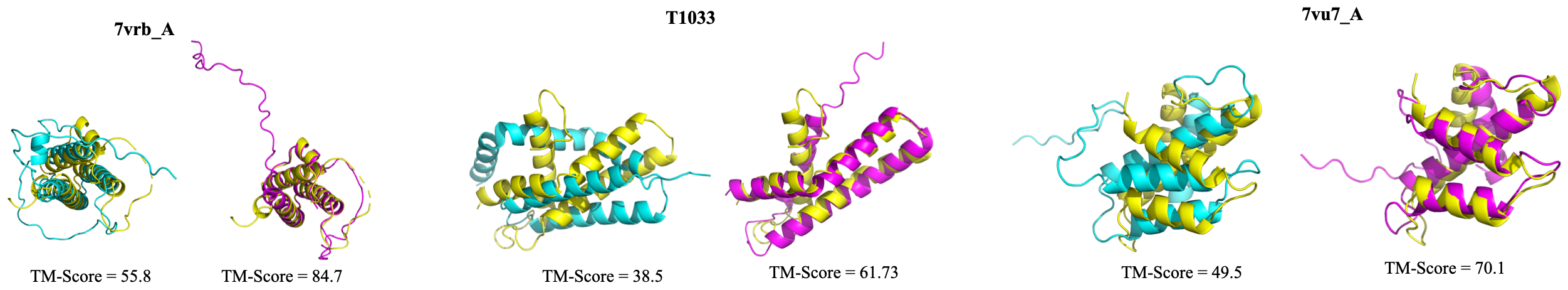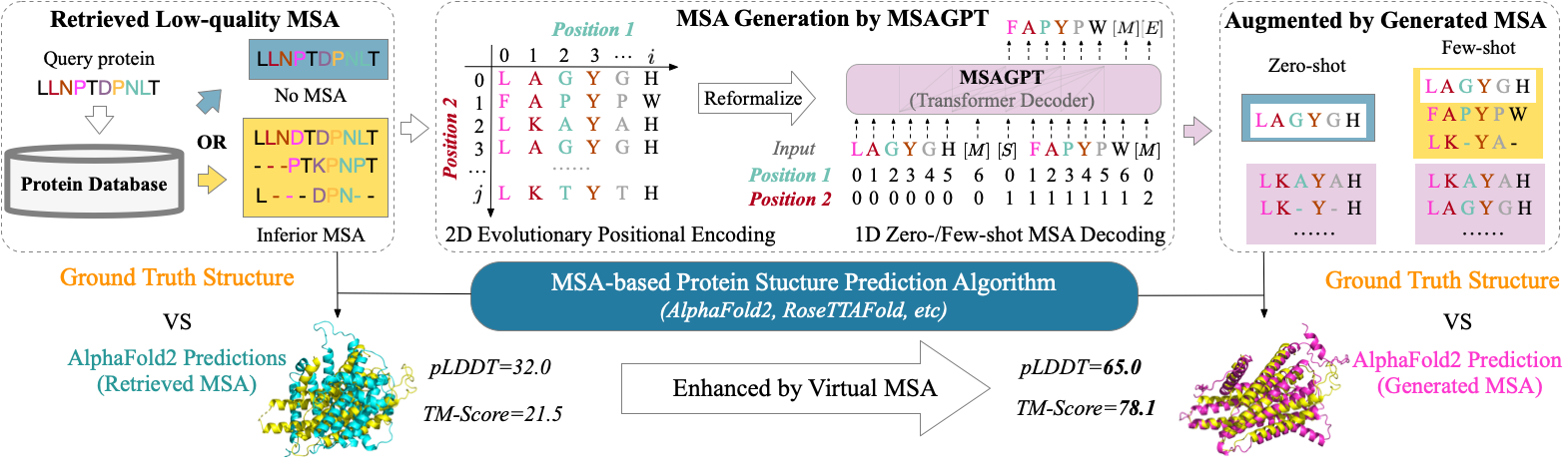
MSAGPT📖 Paper: MSAGPT: Neural Prompting Protein Structure Prediction via MSA Generative Pre-Training MSAGPT is a powerful protein language model (PLM). MSAGPT has 3 billion parameters with three versions of the model, MSAGPT, MSAGPT-Sft, and MSAGPT-Dpo, supporting zero-shot and few-shot MSA generation. MSAGPT achieves state-of-the-art structural prediction performance on natural MSA-scarce scenarios. |