---
license: agpl-3.0
tags:
- image
- keras
- myology
- biology
- histology
- muscle
- cells
- fibers
- myopathy
- SDH
- myoquant
- classification
- mitochondria
datasets:
- corentinm7/MyoQuant-SDH-Data
metrics:
- accuracy
library_name: keras
model-index:
- name: MyoQuant-SDH-Resnet50V2
results:
- task:
type: image-classification # Required. Example: automatic-speech-recognition
name: Image Classification # Optional. Example: Speech Recognition
dataset:
type: corentinm7/MyoQuant-SDH-Data # Required. Example: common_voice. Use dataset id from https://hf.co/datasets
name: MyoQuant SDH Data # Required. A pretty name for the dataset. Example: Common Voice (French)
split: test # Optional. Example: test
metrics:
- type: accuracy # Required. Example: wer. Use metric id from https://hf.co/metrics
value: 0.932 # Required. Example: 20.90
name: Test Accuracy # Optional. Example: Test WER
---
## Model description

This is the model card for the SDH Model used by the [MyoQuant](https://github.com/lambda-science/MyoQuant) tool.
## Intended uses & limitations
It's intended to allow people to use, improve and verify the reproducibility of our MyoQuant tool. The SDH model is used to classify SDH stained muscle fiber with abnormal mitochondria profile.
## Training and evaluation data
It's trained on the [corentinm7/MyoQuant-SDH-Data](https://huggingface.co/datasets/corentinm7/MyoQuant-SDH-Data), avaliable on HuggingFace Dataset Hub.
## Training procedure
This model was trained using the ResNet50V2 model architecture in Keras.
All images have been resized to 256x256 using the `tf.image.resize()` function from Tensorflow.
Data augmentation was included as layers before ResNet50V2.
Full model code:
```python
data_augmentation = tf.keras.Sequential([
layers.RandomBrightness(factor=0.2, input_shape=(None, None, 3)), # Not avaliable in tensorflow 2.8
layers.RandomContrast(factor=0.2),
layers.RandomFlip("horizontal_and_vertical"),
layers.RandomRotation(0.3, fill_mode="constant"),
layers.RandomZoom(.2, .2, fill_mode="constant"),
layers.RandomTranslation(0.2, .2,fill_mode="constant"),
layers.Resizing(256, 256, interpolation="bilinear", crop_to_aspect_ratio=True),
layers.Rescaling(scale=1./127.5, offset=-1), # For [-1, 1] scaling
])
# My ResNet50V2
model = models.Sequential()
model.add(data_augmentation)
model.add(
ResNet50V2(
include_top=False,
input_shape=(256,256,3),
pooling="avg",
)
)
model.add(layers.Flatten())
model.add(layers.Dense(len(config.SUB_FOLDERS), activation='softmax'))
```
```
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
sequential (Sequential) (None, 256, 256, 3) 0
resnet50v2 (Functional) (None, 2048) 23564800
flatten (Flatten) (None, 2048) 0
dense (Dense) (None, 2) 4098
=================================================================
Total params: 23,568,898
Trainable params: 23,523,458
Non-trainable params: 45,440
_________________________________________________________________
```
We used a ResNet50V2 pre-trained on ImageNet as a starting point and trained the model using an EarlyStopping with a value of 20 (i.e. if validation loss doesn't improve after 20 epoch, stop the training and roll back to the epoch with lowest val loss.)
Class imbalance was handled by using the class\_-weight attribute during training. It was calculated for each class as `(1/n. elem of the class) * (n. of all training elem / 2)` giving in our case: `{0: 0.6593016912165849, 1: 2.069349315068493}`
### Training hyperparameters
The following hyperparameters were used during training:
- optimizer: Adam
- Learning Rate Schedule: `ReduceLROnPlateau(monitor='val_loss', factor=0.2, patience=5, min_lr=1e-7` with START_LR = 1e-5 and MIN_LR = 1e-7
- Loss Function: SparseCategoricalCrossentropy
- Metric: Accuracy
For more details please see the training notebook associated.
## Training Curve
Full training results are avaliable on `Weights and Biases` here: [https://api.wandb.ai/links/lambda-science/ka0iw3b6](https://api.wandb.ai/links/lambda-science/ka0iw3b6)
Plot of the accuracy vs epoch and loss vs epoch for training and validation set.
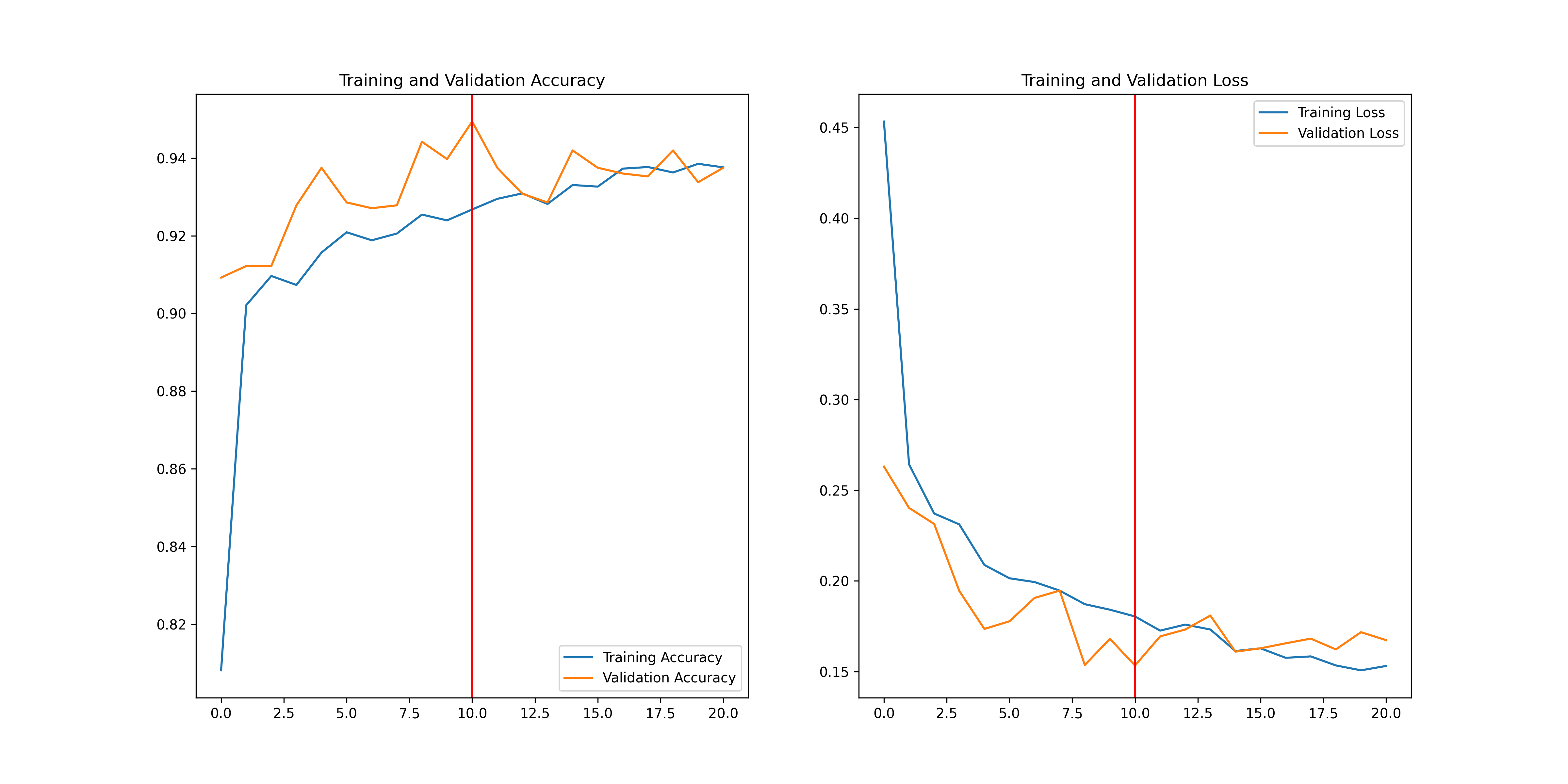
## Test Results
Results for accuracy and balanced accuracy metrics on the test split of the [corentinm7/MyoQuant-SDH-Data](https://huggingface.co/datasets/corentinm7/MyoQuant-SDH-Data) dataset.
```
105/105 - 11s - loss: 0.1574 - accuracy: 0.9321 - 11s/epoch - 102ms/step
Test data results:
0.9321024417877197
105/105 [==============================] - 6s 44ms/step
Test data results:
0.9166411912436779
```
# How to Import the Model
With Tensorflow 2.10 and over:
```python
model_sdh = keras.models.load_model("model.h5")
```
With Tensorflow <2.10:
To import this model RandomBrightness layer had to be added by hand (it was only introduced in Tensorflow 2.10.). So you will need to download the `random_brightness.py` fille in addition to the model.
Then the model can easily be imported in Tensorflow/Keras using:
```python
from .random_brightness import *
model_sdh = keras.models.load_model(
"model.h5", custom_objects={"RandomBrightness": RandomBrightness}
)
```
## The Team Behind this Dataset
**The creator, uploader and main maintainer of this model, associated dataset and MyoQuant is:**
- **[Corentin Meyer, PhD in Biomedical AI](https://cmeyer.fr) Email: Github: [@lambda-science](https://github.com/lambda-science)**
Special thanks to the experts that created the data for the dataset and all the time they spend counting cells :
- **Quentin GIRAUD, PhD Student** @ [Department Translational Medicine, IGBMC, CNRS UMR 7104](https://www.igbmc.fr/en/recherche/teams/pathophysiology-of-neuromuscular-diseases), 1 rue Laurent Fries, 67404 Illkirch, France
- **Charlotte GINESTE, Post-Doc** @ [Department Translational Medicine, IGBMC, CNRS UMR 7104](https://www.igbmc.fr/en/recherche/teams/pathophysiology-of-neuromuscular-diseases), 1 rue Laurent Fries, 67404 Illkirch, France
Last but not least thanks to Bertrand Vernay being at the origin of this project:
- **Bertrand VERNAY, Platform Leader** @ [Light Microscopy Facility, IGBMC, CNRS UMR 7104](https://www.igbmc.fr/en/plateformes-technologiques/photonic-microscopy), 1 rue Laurent Fries, 67404 Illkirch, France
## Partners

MyoQuant-SDH-Model is born within the collaboration between the [CSTB Team @ ICube](https://cstb.icube.unistra.fr/en/index.php/Home) led by Julie D. Thompson, the [Morphological Unit of the Institute of Myology of Paris](https://www.institut-myologie.org/en/recherche-2/neuromuscular-investigation-center/morphological-unit/) led by Teresinha Evangelista, the [imagery platform MyoImage of Center of Research in Myology](https://recherche-myologie.fr/technologies/myoimage/) led by Bruno Cadot, [the photonic microscopy platform of the IGMBC](https://www.igbmc.fr/en/plateformes-technologiques/photonic-microscopy) led by Bertrand Vernay and the [Pathophysiology of neuromuscular diseases team @ IGBMC](https://www.igbmc.fr/en/igbmc/a-propos-de-ligbmc/directory/jocelyn-laporte) led by Jocelyn Laporte


