Commit
•
38e5f8e
1
Parent(s):
28da202
Update README.md
Browse files
README.md
CHANGED
|
@@ -32,7 +32,7 @@ This [repository](https://github.com/PathologyDataScience/BCSS) contains the nec
|
|
| 32 |
|
| 33 |
- Amgad, Mohamed, et al. "NuCLS: A Scalable Crowdsourcing Approach and Dataset for Nucleus Classification and Segmentation in Breast Cancer." GigaScience, vol. 11, 2022, https://doi.org/10.1093/gigascience/giac037. Accessed 18 Mar. 2024.
|
| 34 |
|
| 35 |
-

|
| 36 |
|
| 37 |
|
| 38 |
## Accessing the Data
|
|
@@ -71,7 +71,7 @@ A single dataset entry contains the following details:
|
|
| 71 |
|
| 72 |
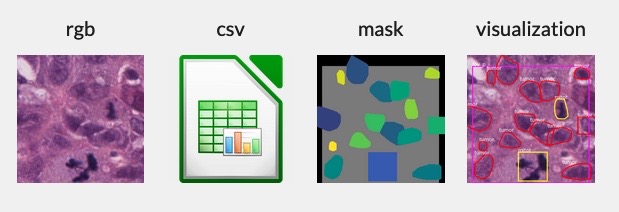
- `mask_image`: A mask image with each nucleus labeled. Class labels are encoded in the first channel. The second and third channels are used to create a unique identifier for each nucleus. The field of view (gray area) is marked to delineate the annotated region.
|
| 73 |
|
| 74 |
-
[This file](https://
|
| 75 |
|
| 76 |
- `visualization_image`: A visualization image that overlays the RGB and mask images to assist in interpretability.
|
| 77 |
|
|
@@ -89,7 +89,7 @@ A single dataset entry contains the following details:
|
|
| 89 |
|
| 90 |
- `coords_x`, `coords_y`: The specific boundary coordinates of the nucleus.
|
| 91 |
|
| 92 |
-

|
| 36 |
|
| 37 |
|
| 38 |
## Accessing the Data
|
|
|
|
| 71 |
|
| 72 |
- `mask_image`: A mask image with each nucleus labeled. Class labels are encoded in the first channel. The second and third channels are used to create a unique identifier for each nucleus. The field of view (gray area) is marked to delineate the annotated region.
|
| 73 |
|
| 74 |
+
[This file](https://huggingface.co/datasets/minhanhto09/NuCLS_dataset/resolve/main/Images/fig2.png) contains the nucleus label encoding, including a special 'fov' code encoding the intended annotation region.
|
| 75 |
|
| 76 |
- `visualization_image`: A visualization image that overlays the RGB and mask images to assist in interpretability.
|
| 77 |
|
|
|
|
| 89 |
|
| 90 |
- `coords_x`, `coords_y`: The specific boundary coordinates of the nucleus.
|
| 91 |
|
| 92 |
+
|
| 93 |
|
| 94 |
### Data Split
|
| 95 |
|
|
|
|
| 115 |
|
| 116 |
- `train_fold_999`: 21 examples
|
| 117 |
- `test_fold_999`: 7 examples
|
| 118 |
+
|
| 119 |
+
Note that the debug configuration utilizes these particular folds `train_fold_999` and `test_fold_999` due to their smaller numbers of examples.
|
| 120 |
|
| 121 |
## Usage Example
|
| 122 |
|