Datasets:
Upload folder using huggingface_hub
Browse files- README.md +206 -0
- data.parquet +3 -0
README.md
ADDED
|
@@ -0,0 +1,206 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
language: rna
|
| 3 |
+
tags:
|
| 4 |
+
- Biology
|
| 5 |
+
- RNA
|
| 6 |
+
license:
|
| 7 |
+
- agpl-3.0
|
| 8 |
+
size_categories:
|
| 9 |
+
- 100K<n<1M
|
| 10 |
+
source_datasets:
|
| 11 |
+
- multimolecule/crw
|
| 12 |
+
- multimolecule/tmrna_website
|
| 13 |
+
- multimolecule/srpdb
|
| 14 |
+
- multimolecule/spr
|
| 15 |
+
- multimolecule/rnp
|
| 16 |
+
- multimolecule/rfam
|
| 17 |
+
- multimolecule/pdb
|
| 18 |
+
task_categories:
|
| 19 |
+
- text-generation
|
| 20 |
+
- fill-mask
|
| 21 |
+
task_ids:
|
| 22 |
+
- language-modeling
|
| 23 |
+
- masked-language-modeling
|
| 24 |
+
pretty_name: bpRNA-1m
|
| 25 |
+
library_name: multimolecule
|
| 26 |
+
---
|
| 27 |
+
|
| 28 |
+
# bpRNA-1m
|
| 29 |
+
|
| 30 |
+
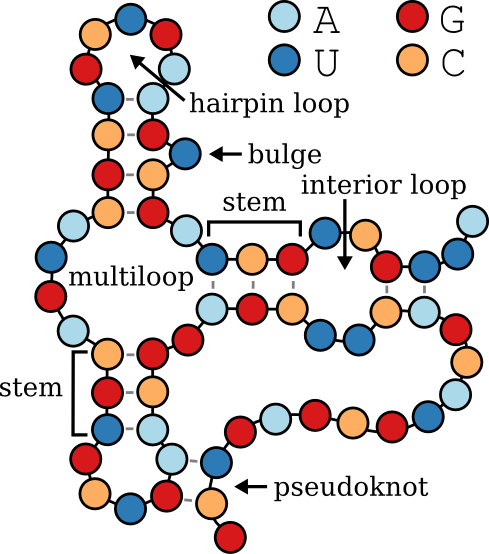
|
| 31 |
+
|
| 32 |
+
bpRNA-1m is a database of single molecule secondary structures annotated using bpRNA.
|
| 33 |
+
|
| 34 |
+
## Disclaimer
|
| 35 |
+
|
| 36 |
+
This is an UNOFFICIAL release of the [bpRNA-1m](https://bprna.cgrb.oregonstate.edu/index.html) by Center for Quantitative Life Sciences of the Oregon State University.
|
| 37 |
+
|
| 38 |
+
**The team releasing bpRNA did not write this dataset card for this dataset so this dataset card has been written by the MultiMolecule team.**
|
| 39 |
+
|
| 40 |
+
## Dataset Description
|
| 41 |
+
|
| 42 |
+
- **Homepage**: https://multimolecule.danling.org/datasets/bprna
|
| 43 |
+
- **datasets**: https://huggingface.co/datasets/multimolecule/bprna
|
| 44 |
+
- **Point of Contact**: [Center for Quantitative Life Sciences of the Oregon State University](https://cqls.oregonstate.edu)
|
| 45 |
+
- **Original URL**: https://bprna.cgrb.oregonstate.edu/index.html
|
| 46 |
+
|
| 47 |
+
## Example Entry
|
| 48 |
+
|
| 49 |
+
| id | sequence | secondary_structure | structural_annotation | functional_annotation |
|
| 50 |
+
| ------------- | ---------------------------------------------- | --------------------------------------------------- | -------------------------------------------------- | ---------------------------------------------- |
|
| 51 |
+
| bpRNA_CRW_170 | GGCUCACCAAGGCGACGACGGGUAGCCGGCCUGAGAGGGCGAC... | ((((((((....))))...))))....((((((..........)))))... | EEEEEEEEEEEEEEEEEEEEEEEESSSSSSSSHHHHSSSSBBBSSSS... | NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN |
|
| 52 |
+
|
| 53 |
+
## Column Description
|
| 54 |
+
|
| 55 |
+
The converted dataset consists of the following columns, each providing specific information about the RNA secondary structures, consistent with the bpRNA standard:
|
| 56 |
+
|
| 57 |
+
- **id**:
|
| 58 |
+
A unique identifier for each RNA entry. This ID is derived from the original `.sta` file name and serves as a reference to the specific RNA structure within the dataset.
|
| 59 |
+
|
| 60 |
+
- **sequence**:
|
| 61 |
+
The nucleotide sequence of the RNA molecule, represented using the standard RNA bases:
|
| 62 |
+
|
| 63 |
+
- **A**: Adenine
|
| 64 |
+
- **C**: Cytosine
|
| 65 |
+
- **G**: Guanine
|
| 66 |
+
- **U**: Uracil
|
| 67 |
+
|
| 68 |
+
- **secondary_structure**:
|
| 69 |
+
The secondary structure of the RNA represented in dot-bracket notation, using up to three types of symbols to indicate base pairing and unpaired regions, as per bpRNA's standard:
|
| 70 |
+
|
| 71 |
+
- **Dots (`.`)**: Represent unpaired nucleotides.
|
| 72 |
+
- **Parentheses (`(` and `)`)**: Represent base pairs in standard stems (page 1).
|
| 73 |
+
- **Square Brackets (`[` and `]`)**: Represent base pairs in pseudoknots (page 2).
|
| 74 |
+
- **Curly Braces (`{` and `}`)**: Represent base pairs in additional pseudoknots (page 3).
|
| 75 |
+
|
| 76 |
+
- **structural_annotation**:
|
| 77 |
+
Structural annotations categorizing different regions of the RNA based on their roles within the secondary structure, consistent with bpRNA standards:
|
| 78 |
+
|
| 79 |
+
- **E**: **External Loop** – Regions that are unpaired and external to any loop or helix.
|
| 80 |
+
- **S**: **Stem** – Paired regions forming helical structures.
|
| 81 |
+
- **H**: **Hairpin Loop** – Unpaired regions at the end of a stem, forming a loop.
|
| 82 |
+
- **I**: **Internal Loop** – Unpaired regions between two stems.
|
| 83 |
+
- **M**: **Multi-loop** – Junctions where three or more stems converge.
|
| 84 |
+
- **B**: **Bulge** – Unpaired nucleotides on one side of a stem.
|
| 85 |
+
- **X**: **Ambiguous** or **Undetermined** – Regions where the structure is unknown or cannot be classified.
|
| 86 |
+
- **K**: **Pseudoknot** – Regions involved in pseudoknots, where base pairs cross each other.
|
| 87 |
+
|
| 88 |
+
- **functional_annotation**:
|
| 89 |
+
Functional annotations indicating specific functional elements or regions within the RNA sequence, as defined by bpRNA:
|
| 90 |
+
- **N**: **None** – No specific functional annotation is assigned.
|
| 91 |
+
- **K**: **Pseudoknot** – Marks nucleotides involved in pseudoknot structures, which can be functionally significant.
|
| 92 |
+
|
| 93 |
+
## Variations
|
| 94 |
+
|
| 95 |
+
This dataset is available in two variants:
|
| 96 |
+
|
| 97 |
+
- [bpRNA-1m](https://huggingface.co/datasets/multimolecule/bprna): The main bpRNA-1m dataset.
|
| 98 |
+
- [bpRNA-1m(90)](https://huggingface.co/datasets/multimolecule/bprna-90): bpRNA_1m(90) is a subset of bpRNA_1m containing RNAs with less than 90% sequence similarity.
|
| 99 |
+
|
| 100 |
+
## License
|
| 101 |
+
|
| 102 |
+
This dataset is licensed under the [AGPL-3.0 License](https://www.gnu.org/licenses/agpl-3.0.html).
|
| 103 |
+
|
| 104 |
+
```spdx
|
| 105 |
+
SPDX-License-Identifier: AGPL-3.0-or-later
|
| 106 |
+
```
|
| 107 |
+
|
| 108 |
+
## Citation
|
| 109 |
+
|
| 110 |
+
```bibtex
|
| 111 |
+
@article{danaee2018bprna,
|
| 112 |
+
author = {Danaee, Padideh and Rouches, Mason and Wiley, Michelle and Deng, Dezhong and Huang, Liang and Hendrix, David},
|
| 113 |
+
journal = {Nucleic Acids Research},
|
| 114 |
+
month = jun,
|
| 115 |
+
number = 11,
|
| 116 |
+
pages = {5381--5394},
|
| 117 |
+
title = {{bpRNA}: large-scale automated annotation and analysis of {RNA} secondary structure},
|
| 118 |
+
volume = 46,
|
| 119 |
+
year = 2018
|
| 120 |
+
}
|
| 121 |
+
|
| 122 |
+
@article{cannone2002comparative,
|
| 123 |
+
author = {Cannone, Jamie J and Subramanian, Sankar and Schnare, Murray N and Collett, James R and D'Souza, Lisa M and Du, Yushi and Feng, Brian and Lin, Nan and Madabusi, Lakshmi V and M{\"u}ller, Kirsten M and Pande, Nupur and Shang, Zhidi and Yu, Nan and Gutell, Robin R},
|
| 124 |
+
copyright = {https://www.springernature.com/gp/researchers/text-and-data-mining},
|
| 125 |
+
journal = {BMC Bioinformatics},
|
| 126 |
+
month = jan,
|
| 127 |
+
number = 1,
|
| 128 |
+
pages = {2},
|
| 129 |
+
publisher = {Springer Science and Business Media LLC},
|
| 130 |
+
title = {The comparative {RNA} web ({CRW}) site: an online database of comparative sequence and structure information for ribosomal, intron, and other {RNAs}},
|
| 131 |
+
volume = 3,
|
| 132 |
+
year = 2002
|
| 133 |
+
}
|
| 134 |
+
|
| 135 |
+
@article{zwieb2003tmrdb,
|
| 136 |
+
author = {Zwieb, Christian and Gorodkin, Jan and Knudsen, Bjarne and Burks, Jody and Wower, Jacek},
|
| 137 |
+
journal = {Nucleic Acids Research},
|
| 138 |
+
month = jan,
|
| 139 |
+
number = 1,
|
| 140 |
+
pages = {446--447},
|
| 141 |
+
publisher = {Oxford University Press (OUP)},
|
| 142 |
+
title = {{tmRDB} ({tmRNA} database)},
|
| 143 |
+
volume = 31,
|
| 144 |
+
year = 2003
|
| 145 |
+
}
|
| 146 |
+
|
| 147 |
+
@article{rosenblad2003srpdb,
|
| 148 |
+
author = {Rosenblad, Magnus Alm and Gorodkin, Jan and Knudsen, Bjarne and Zwieb, Christian and Samuelsson, Tore},
|
| 149 |
+
journal = {Nucleic Acids Research},
|
| 150 |
+
month = jan,
|
| 151 |
+
number = 1,
|
| 152 |
+
pages = {363--364},
|
| 153 |
+
publisher = {Oxford University Press (OUP)},
|
| 154 |
+
title = {{SRPDB}: Signal Recognition Particle Database},
|
| 155 |
+
volume = 31,
|
| 156 |
+
year = 2003
|
| 157 |
+
}
|
| 158 |
+
|
| 159 |
+
@article{sprinzl2005compilation,
|
| 160 |
+
author = {Sprinzl, Mathias and Vassilenko, Konstantin S},
|
| 161 |
+
journal = {Nucleic Acids Research},
|
| 162 |
+
month = jan,
|
| 163 |
+
number = {Database issue},
|
| 164 |
+
pages = {D139--40},
|
| 165 |
+
publisher = {Oxford University Press (OUP)},
|
| 166 |
+
title = {Compilation of {tRNA} sequences and sequences of {tRNA} genes},
|
| 167 |
+
volume = 33,
|
| 168 |
+
year = 2005
|
| 169 |
+
}
|
| 170 |
+
|
| 171 |
+
@article{brown1994ribonuclease,
|
| 172 |
+
author = {Brown, J W and Haas, E S and Gilbert, D G and Pace, N R},
|
| 173 |
+
journal = {Nucleic Acids Research},
|
| 174 |
+
month = sep,
|
| 175 |
+
number = 17,
|
| 176 |
+
pages = {3660--3662},
|
| 177 |
+
publisher = {Oxford University Press (OUP)},
|
| 178 |
+
title = {The Ribonuclease {P} database},
|
| 179 |
+
volume = 22,
|
| 180 |
+
year = 1994
|
| 181 |
+
}
|
| 182 |
+
|
| 183 |
+
@article{griffiths2003rfam,
|
| 184 |
+
author = {Griffiths-Jones, Sam and Bateman, Alex and Marshall, Mhairi and Khanna, Ajay and Eddy, Sean R},
|
| 185 |
+
journal = {Nucleic Acids Research},
|
| 186 |
+
month = jan,
|
| 187 |
+
number = 1,
|
| 188 |
+
pages = {439--441},
|
| 189 |
+
publisher = {Oxford University Press (OUP)},
|
| 190 |
+
title = {Rfam: an {RNA} family database},
|
| 191 |
+
volume = 31,
|
| 192 |
+
year = 2003
|
| 193 |
+
}
|
| 194 |
+
|
| 195 |
+
@article{berman2000protein,
|
| 196 |
+
author = {Berman, H M and Westbrook, J and Feng, Z and Gilliland, G and Bhat, T N and Weissig, H and Shindyalov, I N and Bourne, P E},
|
| 197 |
+
journal = {Nucleic Acids Research},
|
| 198 |
+
month = jan,
|
| 199 |
+
number = 1,
|
| 200 |
+
pages = {235--242},
|
| 201 |
+
publisher = {Oxford University Press (OUP)},
|
| 202 |
+
title = {The Protein Data Bank},
|
| 203 |
+
volume = 28,
|
| 204 |
+
year = 2000
|
| 205 |
+
}
|
| 206 |
+
```
|
data.parquet
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:57e3a9ae6236a96b88dd269c9bcdaaa3655e8012b6f1f04960916896f5fc6c2f
|
| 3 |
+
size 2424694
|