---
license: other
---
# AIDO.Protein2StructureToken-16B
**AIDO.Protein2StructureToken-16B** is a fine-tuned version of [AIDO.Protein-16B](https://huggingface.co/genbio-ai/AIDO.Protein-16B), for protein structure prediction.
This model uses amino acid sequences as input to predict tokens that can be decoded into 3D structures by [AIDO.StructureDecoder](https://huggingface.co/genbio-ai/AIDO.StructureDecoder).
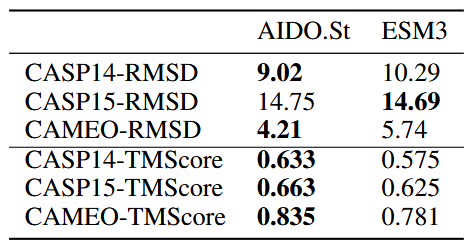
It surpasses existing state-of-the-art models, such as **ESM3-open**, in structure prediction tasks, demonstrating its robustness and capability in this domain.
## Model Architecture Details
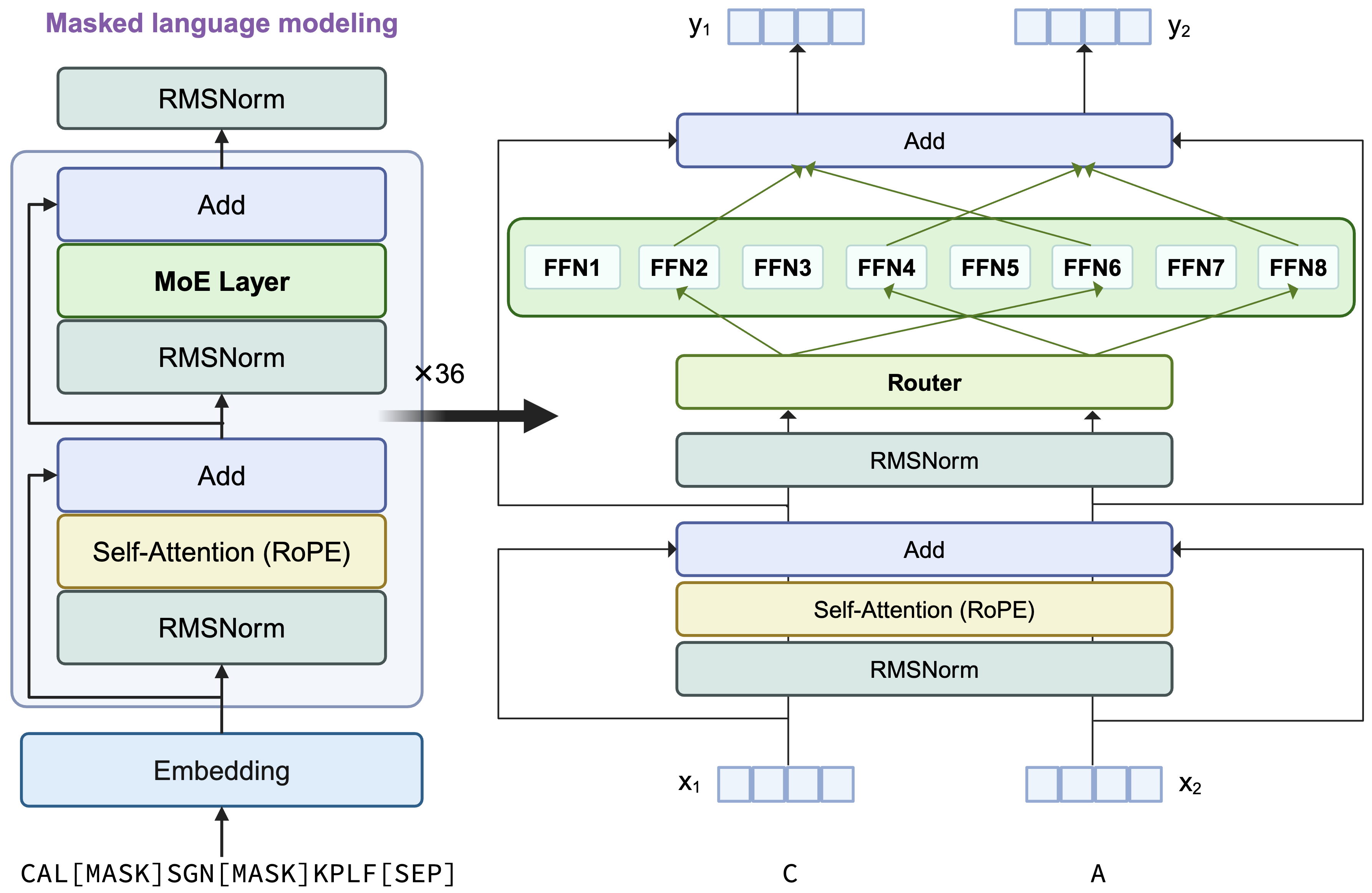
This model retains the architecture of AIDO.Protein-16B, a transformer encoder-only architecture with dense MLP layers replaced by sparse Mixture of Experts (MoE) layers.
Each token activates 2 experts using a top-2 routing mechanism. A visual summary of the architecture is provided below:
 ### Key Differences
The final output linear layer has been adapted to support a new vocabulary size:
- **Input Vocabulary Size**: 44 (amino acids + special tokens)
- **Output Vocabulary Size**: 512 (structure tokens without special tokens)
### Architecture Parameters
| Component | Value |
|-------------------------------|-------|
| Number of Attention Heads | 36 |
| Number of Hidden Layers | 36 |
| Hidden Size | 2304 |
| Number of MoE Layers per Block| 8 |
| Number of MoE Layers per Token| 2 |
| Input Vocabulary Size | 44 |
| Output Vocabulary Size | 512 |
| Context Length | 1024 |
## Training Details
The fine-tuning process used **0.4 trillion tokens**, using AlphaFold database with **170M samples** and PDB database with **0.4M samples**, making it highly specialized for structure prediction. The training took around 20 days on 64 A100 GPUs.
- **Batch Size**: Global batch size of 2048
- **Context Length**: 1024
- **Precision**: FP16
- **Hardware**: 64 NVIDIA A100 80GB GPUs
- **Learning Rate**: Max learning rate of 1e-4
- **Scheduler**: Cosine decay with 2.5% warmup
- **Tokens Trained**: 0.4T tokens
- **Training steps**: 200k steps
## Tokenization
The input sequence should be single-chain amino acid sequences.
- **Input Tokenization**: The sequences are tokenized at the amino acid level and terminated with a `[SEP]` token (id=34).
- **Output Tokenization**: Each input token is converted into a structure token. The output can be decoded into 3D structures in PDB format using [AIDO.StructureDecoder](https://huggingface.co/genbio-ai/AIDO.StructureDecoder).
## Results
### Key Differences
The final output linear layer has been adapted to support a new vocabulary size:
- **Input Vocabulary Size**: 44 (amino acids + special tokens)
- **Output Vocabulary Size**: 512 (structure tokens without special tokens)
### Architecture Parameters
| Component | Value |
|-------------------------------|-------|
| Number of Attention Heads | 36 |
| Number of Hidden Layers | 36 |
| Hidden Size | 2304 |
| Number of MoE Layers per Block| 8 |
| Number of MoE Layers per Token| 2 |
| Input Vocabulary Size | 44 |
| Output Vocabulary Size | 512 |
| Context Length | 1024 |
## Training Details
The fine-tuning process used **0.4 trillion tokens**, using AlphaFold database with **170M samples** and PDB database with **0.4M samples**, making it highly specialized for structure prediction. The training took around 20 days on 64 A100 GPUs.
- **Batch Size**: Global batch size of 2048
- **Context Length**: 1024
- **Precision**: FP16
- **Hardware**: 64 NVIDIA A100 80GB GPUs
- **Learning Rate**: Max learning rate of 1e-4
- **Scheduler**: Cosine decay with 2.5% warmup
- **Tokens Trained**: 0.4T tokens
- **Training steps**: 200k steps
## Tokenization
The input sequence should be single-chain amino acid sequences.
- **Input Tokenization**: The sequences are tokenized at the amino acid level and terminated with a `[SEP]` token (id=34).
- **Output Tokenization**: Each input token is converted into a structure token. The output can be decoded into 3D structures in PDB format using [AIDO.StructureDecoder](https://huggingface.co/genbio-ai/AIDO.StructureDecoder).
## Results
 ## How to Use
### Structure Prediction
To reproduce the structure prediction results described above, follow these steps:
1. Install the [Model Generator package](https://github.com/genbio-ai/ModelGenerator/).
2. Run the prediction command:
```bash
mgen predict --config experiments/AIDO.StructureTokenizer/protein2structoken_16b.yaml
```
This will pull the CASP14, CASP15, and CAMEO dataset from [genbio-ai/casp14-casp15-cameo-test-proteins](https://huggingface.co/datasets/genbio-ai/casp14-casp15-cameo-test-proteins), and predict the structure tokens from the amino acid sequence.
3. Convert the output `.tsv` to `.pt` and extract model codebook:
```bash
# convert the predicted structures in tsv into one pt file
python experiments/AIDO.StructureTokenizer/struct_token_format_conversion.py logs/protein2structoken_16b/predict_predictions.tsv logs/protein2structoken_16b/predict_predictions.pt
# extract the codebook of the structure tokenizer
python experiments/AIDO.StructureTokenizer/extract_structure_tokenizer_codebook.py --output_path logs/protein2structoken_16b/codebook.pt
```
5. Run the decoding command to get 3D structures in PDB format (currently this script only supports single GPU inference):
```bash
CUDA_VISIBLE_DEVICES=0 mgen predict --config experiments/AIDO.StructureTokenizer/decode.yaml \
--data.init_args.config.struct_tokens_datasets_configs.name=protein2structoken_16b \
--data.init_args.config.struct_tokens_datasets_configs.struct_tokens_path=logs/protein2structoken_16b/predict_predictions.pt \
--data.init_args.config.struct_tokens_datasets_configs.codebook_path=logs/protein2structoken_16b/codebook.pt
```
The outputs are in `logs/protstruct_decode/protein2structoken_16b_pdb_files/`
6. You can compare the predicted structures with the ground truth PDBs in [genbio-ai/casp14-casp15-cameo-test-proteins](https://huggingface.co/datasets/genbio-ai/casp14-casp15-cameo-test-proteins/tree/main).
Alternatively, you can provide your own input amino acid sequence in a CSV file. Here is one example csv at `experiments/AIDO.StructureTokenizer/protein2structoken_example_input.csv` in `ModelGenerator`:
```
idx,aa_seq
example,KEFWNLDKNLQLRLGIVFLG
```
Here, `idx` is a unique name, and `aa_seq` is the amino acid sequence. To use this customized CSV file, replace the second step with
```bash
mgen predict --config experiments/AIDO.StructureTokenizer/protein2structoken_16b.yaml \
--data.init_args.path=experiments/AIDO.StructureTokenizer/ \
--data.init_args.test_split_files=[protein2structoken_example_input.csv]
```
### Build any downstream models from this backbone with ModelGenerator
For more information, visit: [Model Generator](https://github.com/genbio-ai/modelgenerator)
```bash
mgen fit --model SequenceClassification --model.backbone aido_protein_16b --data SequenceClassificationDataModule --data.path
mgen test --model SequenceClassification --model.backbone aido_protein_16b --data SequenceClassificationDataModule --data.path
```
The usage of this model is the same as [AIDO.Protein-16B](https://huggingface.co/genbio-ai/AIDO.Protein-16B).
You only need to change the `model.backbone` to `aido_protein2structoken`.
### Or use directly in Python
#### Embedding
```python
from modelgenerator.tasks import Embed
model = Embed.from_config({"model.backbone": "aido_protein2structoken_16b"}).eval()
collated_batch = model.collate({"sequences": ["HELLQ", "WRLD"]})
embedding = model(collated_batch)
print(embedding.shape)
print(embedding)
```
#### Sequence Level Classification
```python
import torch
from modelgenerator.tasks import SequenceClassification
model = SequenceClassification.from_config({"model.backbone": "aido_protein2structoken_16b", "model.n_classes": 2}).eval()
collated_batch = model.collate({"sequences": ["HELLQ", "WRLD"]})
logits = model(collated_batch)
print(logits)
print(torch.argmax(logits, dim=-1))
```
#### Token Level Classification
```python
import torch
from modelgenerator.tasks import TokenClassification
model = TokenClassification.from_config({"model.backbone": "aido_protein2structoken_16b", "model.n_classes": 3}).eval()
collated_batch = model.collate({"sequences": ["HELLQ", "WRLD"]})
logits = model(collated_batch)
print(logits)
print(torch.argmax(logits, dim=-1))
```
#### Regression
```python
from modelgenerator.tasks import SequenceRegression
model = SequenceRegression.from_config({"model.backbone": "aido_protein2structoken_16b"}).eval()
collated_batch = model.collate({"sequences": ["HELLQ", "WRLD"]})
logits = model(collated_batch)
print(logits)
```
## Citation
Please cite AIDO.Protein and AIDO.StructureTokenizer using the following BibTex codes:
```
@inproceedings{zhang_balancing_2024,
title = {Balancing Locality and Reconstruction in Protein Structure Tokenizer},
url = {https://www.biorxiv.org/content/10.1101/2024.12.02.626366v2},
doi = {10.1101/2024.12.02.626366},
publisher = {bioRxiv},
author = {Zhang, Jiayou and Meynard-Piganeau, Barthelemy and Gong, James and Cheng, Xingyi and Luo, Yingtao and Ly, Hugo and Song, Le and Xing, Eric},
year = {2024},
booktitle={NeurIPS 2024 Workshop on Machine Learning in Structural Biology (MLSB)},
}
@inproceedings{sun_mixture_2024,
title = {Mixture of Experts Enable Efficient and Effective Protein Understanding and Design},
url = {https://www.biorxiv.org/content/10.1101/2024.11.29.625425v1},
doi = {10.1101/2024.11.29.625425},
publisher = {bioRxiv},
author = {Sun, Ning and Zou, Shuxian and Tao, Tianhua and Mahbub, Sazan and Li, Dian and Zhuang, Yonghao and Wang, Hongyi and Cheng, Xingyi and Song, Le and Xing, Eric P.},
year = {2024},
booktitle={NeurIPS 2024 Workshop on AI for New Drug Modalities},
}
```
## How to Use
### Structure Prediction
To reproduce the structure prediction results described above, follow these steps:
1. Install the [Model Generator package](https://github.com/genbio-ai/ModelGenerator/).
2. Run the prediction command:
```bash
mgen predict --config experiments/AIDO.StructureTokenizer/protein2structoken_16b.yaml
```
This will pull the CASP14, CASP15, and CAMEO dataset from [genbio-ai/casp14-casp15-cameo-test-proteins](https://huggingface.co/datasets/genbio-ai/casp14-casp15-cameo-test-proteins), and predict the structure tokens from the amino acid sequence.
3. Convert the output `.tsv` to `.pt` and extract model codebook:
```bash
# convert the predicted structures in tsv into one pt file
python experiments/AIDO.StructureTokenizer/struct_token_format_conversion.py logs/protein2structoken_16b/predict_predictions.tsv logs/protein2structoken_16b/predict_predictions.pt
# extract the codebook of the structure tokenizer
python experiments/AIDO.StructureTokenizer/extract_structure_tokenizer_codebook.py --output_path logs/protein2structoken_16b/codebook.pt
```
5. Run the decoding command to get 3D structures in PDB format (currently this script only supports single GPU inference):
```bash
CUDA_VISIBLE_DEVICES=0 mgen predict --config experiments/AIDO.StructureTokenizer/decode.yaml \
--data.init_args.config.struct_tokens_datasets_configs.name=protein2structoken_16b \
--data.init_args.config.struct_tokens_datasets_configs.struct_tokens_path=logs/protein2structoken_16b/predict_predictions.pt \
--data.init_args.config.struct_tokens_datasets_configs.codebook_path=logs/protein2structoken_16b/codebook.pt
```
The outputs are in `logs/protstruct_decode/protein2structoken_16b_pdb_files/`
6. You can compare the predicted structures with the ground truth PDBs in [genbio-ai/casp14-casp15-cameo-test-proteins](https://huggingface.co/datasets/genbio-ai/casp14-casp15-cameo-test-proteins/tree/main).
Alternatively, you can provide your own input amino acid sequence in a CSV file. Here is one example csv at `experiments/AIDO.StructureTokenizer/protein2structoken_example_input.csv` in `ModelGenerator`:
```
idx,aa_seq
example,KEFWNLDKNLQLRLGIVFLG
```
Here, `idx` is a unique name, and `aa_seq` is the amino acid sequence. To use this customized CSV file, replace the second step with
```bash
mgen predict --config experiments/AIDO.StructureTokenizer/protein2structoken_16b.yaml \
--data.init_args.path=experiments/AIDO.StructureTokenizer/ \
--data.init_args.test_split_files=[protein2structoken_example_input.csv]
```
### Build any downstream models from this backbone with ModelGenerator
For more information, visit: [Model Generator](https://github.com/genbio-ai/modelgenerator)
```bash
mgen fit --model SequenceClassification --model.backbone aido_protein_16b --data SequenceClassificationDataModule --data.path
mgen test --model SequenceClassification --model.backbone aido_protein_16b --data SequenceClassificationDataModule --data.path
```
The usage of this model is the same as [AIDO.Protein-16B](https://huggingface.co/genbio-ai/AIDO.Protein-16B).
You only need to change the `model.backbone` to `aido_protein2structoken`.
### Or use directly in Python
#### Embedding
```python
from modelgenerator.tasks import Embed
model = Embed.from_config({"model.backbone": "aido_protein2structoken_16b"}).eval()
collated_batch = model.collate({"sequences": ["HELLQ", "WRLD"]})
embedding = model(collated_batch)
print(embedding.shape)
print(embedding)
```
#### Sequence Level Classification
```python
import torch
from modelgenerator.tasks import SequenceClassification
model = SequenceClassification.from_config({"model.backbone": "aido_protein2structoken_16b", "model.n_classes": 2}).eval()
collated_batch = model.collate({"sequences": ["HELLQ", "WRLD"]})
logits = model(collated_batch)
print(logits)
print(torch.argmax(logits, dim=-1))
```
#### Token Level Classification
```python
import torch
from modelgenerator.tasks import TokenClassification
model = TokenClassification.from_config({"model.backbone": "aido_protein2structoken_16b", "model.n_classes": 3}).eval()
collated_batch = model.collate({"sequences": ["HELLQ", "WRLD"]})
logits = model(collated_batch)
print(logits)
print(torch.argmax(logits, dim=-1))
```
#### Regression
```python
from modelgenerator.tasks import SequenceRegression
model = SequenceRegression.from_config({"model.backbone": "aido_protein2structoken_16b"}).eval()
collated_batch = model.collate({"sequences": ["HELLQ", "WRLD"]})
logits = model(collated_batch)
print(logits)
```
## Citation
Please cite AIDO.Protein and AIDO.StructureTokenizer using the following BibTex codes:
```
@inproceedings{zhang_balancing_2024,
title = {Balancing Locality and Reconstruction in Protein Structure Tokenizer},
url = {https://www.biorxiv.org/content/10.1101/2024.12.02.626366v2},
doi = {10.1101/2024.12.02.626366},
publisher = {bioRxiv},
author = {Zhang, Jiayou and Meynard-Piganeau, Barthelemy and Gong, James and Cheng, Xingyi and Luo, Yingtao and Ly, Hugo and Song, Le and Xing, Eric},
year = {2024},
booktitle={NeurIPS 2024 Workshop on Machine Learning in Structural Biology (MLSB)},
}
@inproceedings{sun_mixture_2024,
title = {Mixture of Experts Enable Efficient and Effective Protein Understanding and Design},
url = {https://www.biorxiv.org/content/10.1101/2024.11.29.625425v1},
doi = {10.1101/2024.11.29.625425},
publisher = {bioRxiv},
author = {Sun, Ning and Zou, Shuxian and Tao, Tianhua and Mahbub, Sazan and Li, Dian and Zhuang, Yonghao and Wang, Hongyi and Cheng, Xingyi and Song, Le and Xing, Eric P.},
year = {2024},
booktitle={NeurIPS 2024 Workshop on AI for New Drug Modalities},
}
```
