Push model using huggingface_hub.
Browse files
README.md
CHANGED
|
@@ -16,15 +16,15 @@ tags:
|
|
| 16 |
|
| 17 |
- **Developers:** IBM Research
|
| 18 |
- **GitHub Repository:** [https://github.com/BiomedSciAI/biomed-multi-view](https://github.com/BiomedSciAI/biomed-multi-view)
|
| 19 |
-
- **Paper:** [Multi-view biomedical foundation models for molecule-target and property prediction](https://arxiv.org/abs/
|
| 20 |
-
- **Release Date**: Oct
|
| 21 |
-
- **License:** [
|
| 22 |
|
| 23 |
## Model Description
|
| 24 |
|
| 25 |
-
|
| 26 |
|
| 27 |
-
|
| 28 |
|
| 29 |
|
| 30 |
|
|
@@ -34,13 +34,19 @@ It was introduced in the paper [Multi-view biomedical foundation models for mole
|
|
| 34 |
|
| 35 |
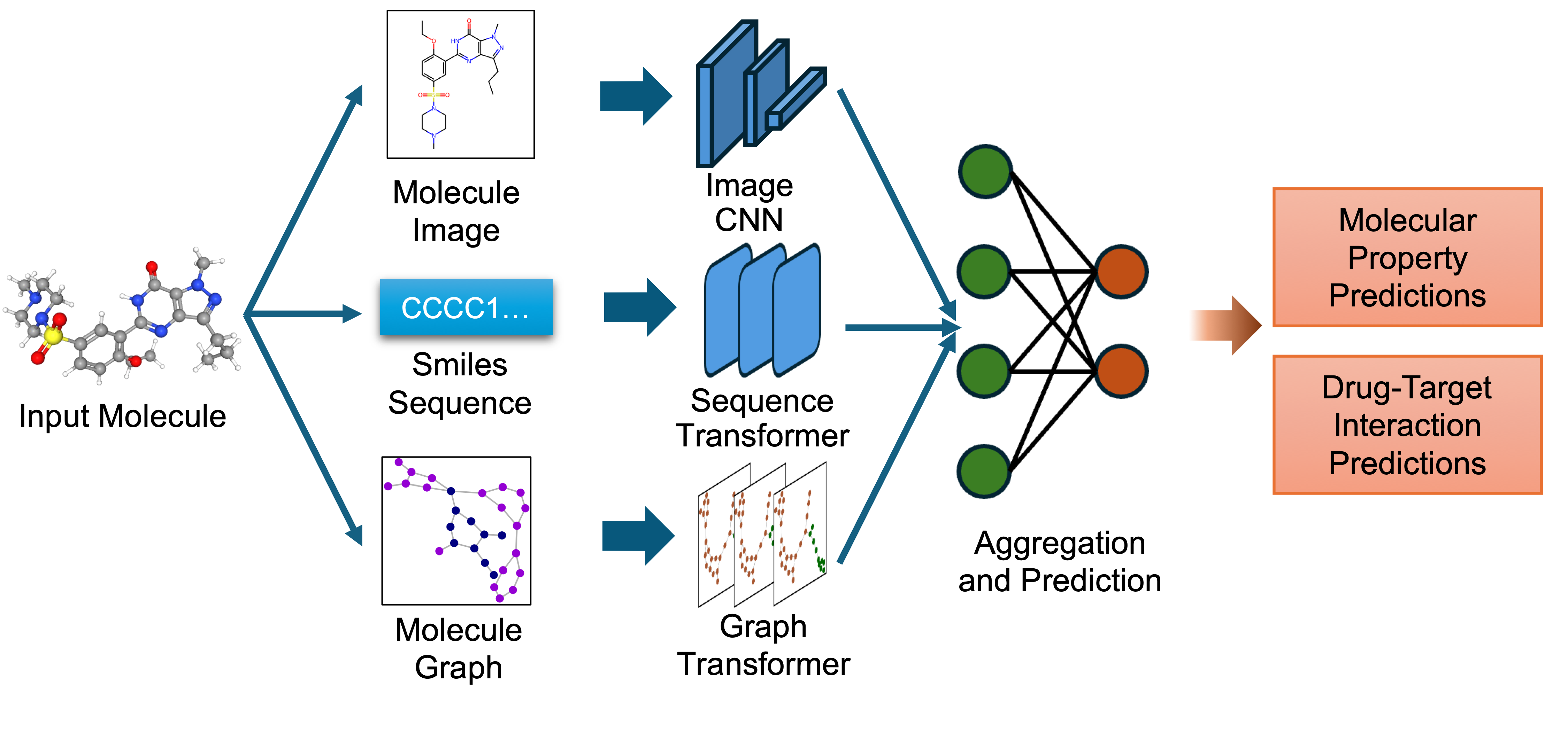
The embeddings from these single-view pre-trained encoders are combined using an attention-based aggregator module. This module learns to weight each view appropriately, producing a unified multi-view embedding. This approach leverages the strengths of each representation to improve performance on downstream predictive tasks.
|
| 36 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 37 |
|
| 38 |
## Usage
|
| 39 |
|
| 40 |
-
Using `SmallMoleculeMultiView` requires [https://github.com/BiomedSciAI/biomed-multi-view](https://github.com/BiomedSciAI/biomed-multi-view)
|
| 41 |
|
| 42 |
## Installation
|
| 43 |
-
Follow these steps to set up the `biomed
|
| 44 |
|
| 45 |
### Prerequisites
|
| 46 |
* Operating System: Linux or macOS
|
|
@@ -50,27 +56,14 @@ Follow these steps to set up the `biomed.multi-view` codebase on your system.
|
|
| 50 |
|
| 51 |
|
| 52 |
### Step 1: Set up the project directory
|
| 53 |
-
Choose a root directory where you want to install biomed
|
| 54 |
|
| 55 |
```bash
|
| 56 |
export ROOT_DIR=~/biomed-multiview
|
| 57 |
mkdir -p $ROOT_DIR
|
| 58 |
```
|
| 59 |
|
| 60 |
-
|
| 61 |
-
If you have Anconda in your system you can skip this step.
|
| 62 |
-
``` bash
|
| 63 |
-
cd $ROOT_DIR
|
| 64 |
-
# Download the Anaconda installer
|
| 65 |
-
wget https://repo.anaconda.com/archive/Anaconda3-2023.03-Linux-x86_64.sh
|
| 66 |
-
|
| 67 |
-
# Run the installer
|
| 68 |
-
bash Anaconda3-2023.03-Linux-x86_64.sh
|
| 69 |
-
# After installation, initialize Conda:
|
| 70 |
-
source activate $ROOT_DIR/anaconda3/bin/activate
|
| 71 |
-
```
|
| 72 |
-
|
| 73 |
-
#### Step 3: Create and activate a Conda environment
|
| 74 |
```bash
|
| 75 |
conda create -y python=3.11 --prefix $ROOT_DIR/envs/biomed-multiview
|
| 76 |
```
|
|
@@ -79,7 +72,7 @@ Activate the environment:
|
|
| 79 |
conda activate $ROOT_DIR/envs/biomed-multiview
|
| 80 |
```
|
| 81 |
|
| 82 |
-
#### Step
|
| 83 |
Navigate to the project directory and clone the repository:
|
| 84 |
```bash
|
| 85 |
mkdir -p $ROOT_DIR/code
|
|
@@ -89,14 +82,14 @@ cd $ROOT_DIR/code
|
|
| 89 |
git clone https://github.com/BiomedSciAI/biomed-multi-view.git
|
| 90 |
|
| 91 |
# Navigate into the cloned repository
|
| 92 |
-
cd biomed
|
| 93 |
```
|
| 94 |
Note: If you prefer using SSH, ensure that your SSH keys are set up with GitHub and use the following command:
|
| 95 |
```bash
|
| 96 |
git clone git@github.com:BiomedSciAI/biomed-multi-view.git
|
| 97 |
```
|
| 98 |
|
| 99 |
-
#### Step
|
| 100 |
Install the package in editable mode along with development dependencies:
|
| 101 |
``` bash
|
| 102 |
pip install -e .['dev']
|
|
@@ -106,7 +99,7 @@ Install additional requirements:
|
|
| 106 |
pip install -r requirements.txt
|
| 107 |
```
|
| 108 |
|
| 109 |
-
#### Step
|
| 110 |
If you are using a Mac with Apple Silicon (M1/M2/M3) and the zsh shell, you may need to disable globbing for the installation command:
|
| 111 |
|
| 112 |
``` bash
|
|
@@ -117,7 +110,7 @@ Install macOS-specific requirements optimized for Apple’s Metal Performance Sh
|
|
| 117 |
pip install -r requirements-mps.txt
|
| 118 |
```
|
| 119 |
|
| 120 |
-
#### Step
|
| 121 |
Verify that the installation was successful by running unit tests
|
| 122 |
|
| 123 |
```bash
|
|
@@ -127,7 +120,8 @@ python -m unittest bmfm_sm.tests.all_tests
|
|
| 127 |
|
| 128 |
### Get embedding example
|
| 129 |
|
| 130 |
-
|
|
|
|
| 131 |
```python
|
| 132 |
# Necessary imports
|
| 133 |
from bmfm_sm.api.smmv_api import SmallMoleculeMultiViewModel
|
|
@@ -152,6 +146,8 @@ print(example_emb.shape)
|
|
| 152 |
|
| 153 |
### Get prediction example
|
| 154 |
|
|
|
|
|
|
|
| 155 |
``` python
|
| 156 |
from bmfm_sm.api.smmv_api import SmallMoleculeMultiViewModel
|
| 157 |
from bmfm_sm.api.dataset_registry import DatasetRegistry
|
|
@@ -160,7 +156,7 @@ from bmfm_sm.api.dataset_registry import DatasetRegistry
|
|
| 160 |
dataset_registry = DatasetRegistry()
|
| 161 |
|
| 162 |
# Example SMILES string
|
| 163 |
-
example_smiles = "CC(C)
|
| 164 |
|
| 165 |
# Get dataset information for dataset
|
| 166 |
ds = dataset_registry.get_dataset_info("LIPOPHILICITY")
|
|
@@ -183,7 +179,7 @@ print("Prediction:", prediction)
|
|
| 183 |
|
| 184 |
##### Output:
|
| 185 |
```bash
|
| 186 |
-
Prediction: {'prediction': [
|
| 187 |
```
|
| 188 |
|
| 189 |
For more advanced usage, see our detailed examples at: https://github.com/BiomedSciAI/biomed-multi-view
|
|
@@ -191,12 +187,15 @@ For more advanced usage, see our detailed examples at: https://github.com/Biomed
|
|
| 191 |
|
| 192 |
## Citation
|
| 193 |
|
| 194 |
-
If you found our work useful, please consider
|
| 195 |
```
|
| 196 |
-
@
|
| 197 |
-
|
| 198 |
-
|
| 199 |
-
|
| 200 |
-
|
|
|
|
|
|
|
|
|
|
| 201 |
}
|
| 202 |
```
|
|
|
|
| 16 |
|
| 17 |
- **Developers:** IBM Research
|
| 18 |
- **GitHub Repository:** [https://github.com/BiomedSciAI/biomed-multi-view](https://github.com/BiomedSciAI/biomed-multi-view)
|
| 19 |
+
- **Paper:** [Multi-view biomedical foundation models for molecule-target and property prediction](https://arxiv.org/abs/2410.19704)
|
| 20 |
+
- **Release Date**: Oct 28th, 2024
|
| 21 |
+
- **License:** [Apache 2.0](https://www.apache.org/licenses/LICENSE-2.0)
|
| 22 |
|
| 23 |
## Model Description
|
| 24 |
|
| 25 |
+
`biomed.sm.mv-te-84m` is a biomedical foundation model for small molecules created using MMELON (Multi-view Molecular Embedding with Late Fusion), a flexible approach to aggregate multiple views (sequence, image, graph) of molecules in a foundation model setting. While models based on single view representation typically performs well on some downstream tasks and not others, the multi-view model performs robustly across a wide range of property prediction tasks encompassing ligand-protein binding, molecular solubility, metabolism and toxicity. It has been applied to screen compounds against a large (> 100 targets) set of G Protein-Coupled receptors (GPCRs) to identify strong binders for 33 targets related to Alzheimer’s disease, which are validated through structure-based modeling and identification of key binding motifs [Multi-view biomedical foundation models for molecule-target and property prediction](https://arxiv.org/abs/2410.19704).
|
| 26 |
|
| 27 |
+
Source code is made available in [this repository](https://github.com/BiomedSciAI/biomed-multi-view).
|
| 28 |
|
| 29 |

|
| 30 |
|
|
|
|
| 34 |
|
| 35 |
The embeddings from these single-view pre-trained encoders are combined using an attention-based aggregator module. This module learns to weight each view appropriately, producing a unified multi-view embedding. This approach leverages the strengths of each representation to improve performance on downstream predictive tasks.
|
| 36 |
|
| 37 |
+
## Intended Use and Limitations
|
| 38 |
+
|
| 39 |
+
The model is intended for (1) Molecular property prediction. The pre-trained model may be fine-tuned for both regression and classification tasks. Examples include but are not limited to binding affinity, solubility and toxicity. (2) Pre-trained model embeddings may be used as the basis for similarity measures to search a chemical library. (3) Small molecule embeddings provided by the model may be combined with protein embeddings to fine-tune on tasks that utilize both small molecule and protein representation. (4) Select task-specific fine-tuned models are given as examples. Through listed activities, model may aid in aspects of the molecular discovery such as lead finding or optimization.
|
| 40 |
+
|
| 41 |
+
|
| 42 |
+
The model’s domain of applicability is small, drug-like molecules. It intended for use with molecules less than 1000 Da molecular weight. The MMELON approach itself may be extended to include proteins and other macromolecules but does not at present provide embeddings for such entities. The model is at present not intended for molecular generation. Molecules must be given as a valid SMILES string that represents a valid chemically bonded graph. Invalid inputs will impact performance or lead to error.
|
| 43 |
|
| 44 |
## Usage
|
| 45 |
|
| 46 |
+
Using `SmallMoleculeMultiView` API requires the codebase [https://github.com/BiomedSciAI/biomed-multi-view](https://github.com/BiomedSciAI/biomed-multi-view)
|
| 47 |
|
| 48 |
## Installation
|
| 49 |
+
Follow these steps to set up the `biomed-multi-view` codebase on your system.
|
| 50 |
|
| 51 |
### Prerequisites
|
| 52 |
* Operating System: Linux or macOS
|
|
|
|
| 56 |
|
| 57 |
|
| 58 |
### Step 1: Set up the project directory
|
| 59 |
+
Choose a root directory where you want to install `biomed-multi-view`. For example:
|
| 60 |
|
| 61 |
```bash
|
| 62 |
export ROOT_DIR=~/biomed-multiview
|
| 63 |
mkdir -p $ROOT_DIR
|
| 64 |
```
|
| 65 |
|
| 66 |
+
#### Step 2: Create and activate a Conda environment
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 67 |
```bash
|
| 68 |
conda create -y python=3.11 --prefix $ROOT_DIR/envs/biomed-multiview
|
| 69 |
```
|
|
|
|
| 72 |
conda activate $ROOT_DIR/envs/biomed-multiview
|
| 73 |
```
|
| 74 |
|
| 75 |
+
#### Step 3: Clone the repository
|
| 76 |
Navigate to the project directory and clone the repository:
|
| 77 |
```bash
|
| 78 |
mkdir -p $ROOT_DIR/code
|
|
|
|
| 82 |
git clone https://github.com/BiomedSciAI/biomed-multi-view.git
|
| 83 |
|
| 84 |
# Navigate into the cloned repository
|
| 85 |
+
cd biomed-multi-view
|
| 86 |
```
|
| 87 |
Note: If you prefer using SSH, ensure that your SSH keys are set up with GitHub and use the following command:
|
| 88 |
```bash
|
| 89 |
git clone git@github.com:BiomedSciAI/biomed-multi-view.git
|
| 90 |
```
|
| 91 |
|
| 92 |
+
#### Step 4: Install package dependencies
|
| 93 |
Install the package in editable mode along with development dependencies:
|
| 94 |
``` bash
|
| 95 |
pip install -e .['dev']
|
|
|
|
| 99 |
pip install -r requirements.txt
|
| 100 |
```
|
| 101 |
|
| 102 |
+
#### Step 5: macOS-Specific instructions (Apple Silicon)
|
| 103 |
If you are using a Mac with Apple Silicon (M1/M2/M3) and the zsh shell, you may need to disable globbing for the installation command:
|
| 104 |
|
| 105 |
``` bash
|
|
|
|
| 110 |
pip install -r requirements-mps.txt
|
| 111 |
```
|
| 112 |
|
| 113 |
+
#### Step 6: Installation verification (optional)
|
| 114 |
Verify that the installation was successful by running unit tests
|
| 115 |
|
| 116 |
```bash
|
|
|
|
| 120 |
|
| 121 |
### Get embedding example
|
| 122 |
|
| 123 |
+
You can generate embeddings for a given molecule using the pretrained model with the following code.
|
| 124 |
+
|
| 125 |
```python
|
| 126 |
# Necessary imports
|
| 127 |
from bmfm_sm.api.smmv_api import SmallMoleculeMultiViewModel
|
|
|
|
| 146 |
|
| 147 |
### Get prediction example
|
| 148 |
|
| 149 |
+
You can use the finetuned models to make predictions on new data.
|
| 150 |
+
|
| 151 |
``` python
|
| 152 |
from bmfm_sm.api.smmv_api import SmallMoleculeMultiViewModel
|
| 153 |
from bmfm_sm.api.dataset_registry import DatasetRegistry
|
|
|
|
| 156 |
dataset_registry = DatasetRegistry()
|
| 157 |
|
| 158 |
# Example SMILES string
|
| 159 |
+
example_smiles = "CC(C)C1CCC(C)CC1O"
|
| 160 |
|
| 161 |
# Get dataset information for dataset
|
| 162 |
ds = dataset_registry.get_dataset_info("LIPOPHILICITY")
|
|
|
|
| 179 |
|
| 180 |
##### Output:
|
| 181 |
```bash
|
| 182 |
+
Prediction: {'prediction': [-2.53]}
|
| 183 |
```
|
| 184 |
|
| 185 |
For more advanced usage, see our detailed examples at: https://github.com/BiomedSciAI/biomed-multi-view
|
|
|
|
| 187 |
|
| 188 |
## Citation
|
| 189 |
|
| 190 |
+
If you found our work useful, please consider giving a star to the repo and cite our paper:
|
| 191 |
```
|
| 192 |
+
@misc{suryanarayanan2024multiviewbiomedicalfoundationmodels,
|
| 193 |
+
title={Multi-view biomedical foundation models for molecule-target and property prediction},
|
| 194 |
+
author={Parthasarathy Suryanarayanan and Yunguang Qiu and Shreyans Sethi and Diwakar Mahajan and Hongyang Li and Yuxin Yang and Elif Eyigoz and Aldo Guzman Saenz and Daniel E. Platt and Timothy H. Rumbell and Kenney Ng and Sanjoy Dey and Myson Burch and Bum Chul Kwon and Pablo Meyer and Feixiong Cheng and Jianying Hu and Joseph A. Morrone},
|
| 195 |
+
year={2024},
|
| 196 |
+
eprint={2410.19704},
|
| 197 |
+
archivePrefix={arXiv},
|
| 198 |
+
primaryClass={q-bio.BM},
|
| 199 |
+
url={https://arxiv.org/abs/2410.19704},
|
| 200 |
}
|
| 201 |
```
|