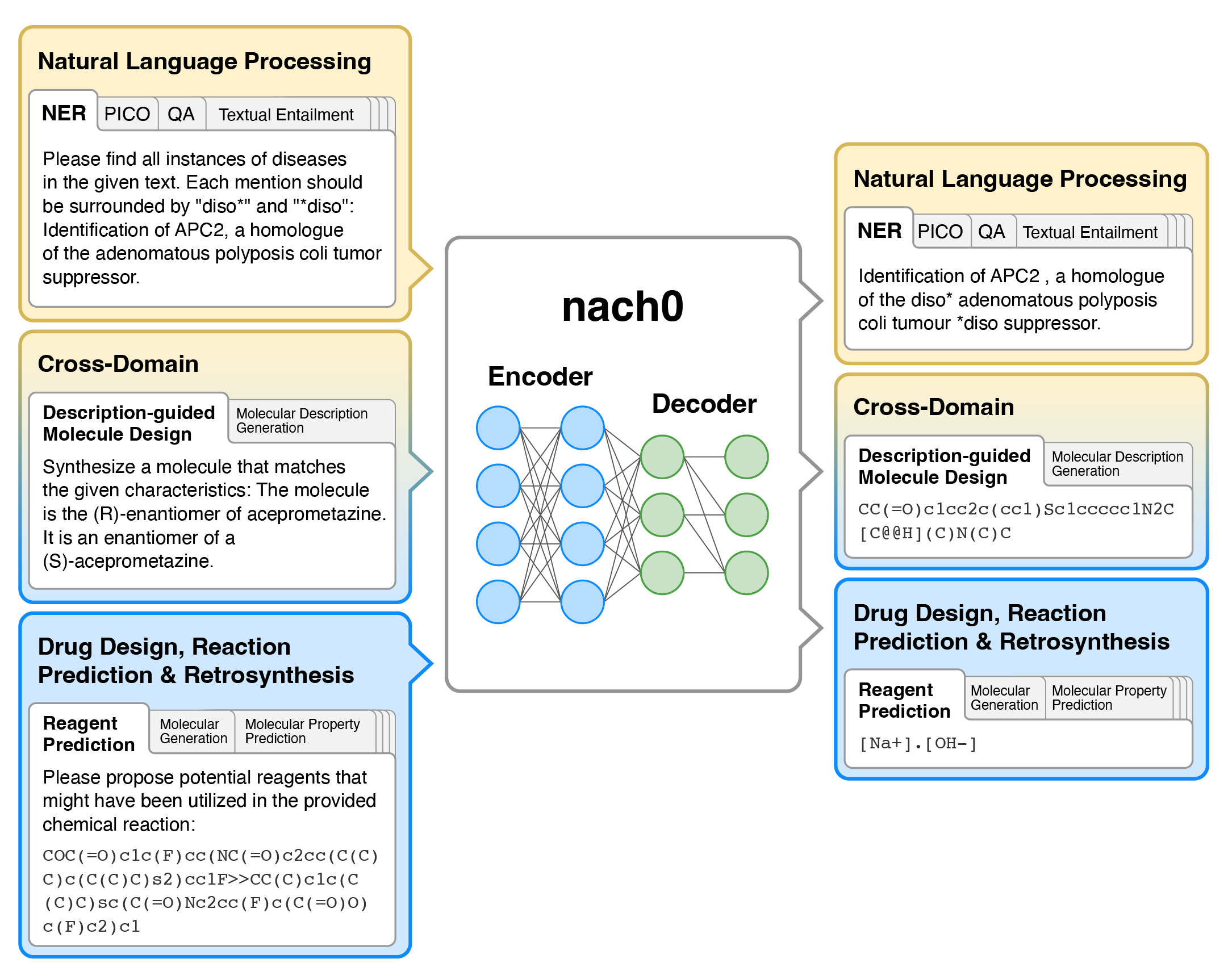
---
license: cc-by-nc-4.0
language:
- en
tags:
- chemistry
---
nach0
Multimodal Natural and Chemical Languages Foundation Model
📃 Paper • ⏬ Base nach0 • ⏬ Large nach0
Overview
- nach0 is a multi-domain and multi-task encoder-decoder LLM pre-trained on unlabeled text from scientific literature, patents, and molecule strings to incorporate a range of chemical and linguistic knowledge.
- We employed instruction tuning, where specific task-related instructions are utilized to fine-tune nach0 for the final set of tasks. To train nach0 effectively, we leverage the NeMo framework, enabling efficient parallel optimization of both base and large model versions.
- Extensive experiments demonstrate that our model outperforms state-of-the-art baselines on single-domain and cross-domain tasks. Furthermore, it can generate high-quality outputs in molecular and textual formats, showcasing its effectiveness in multi-domain setups.
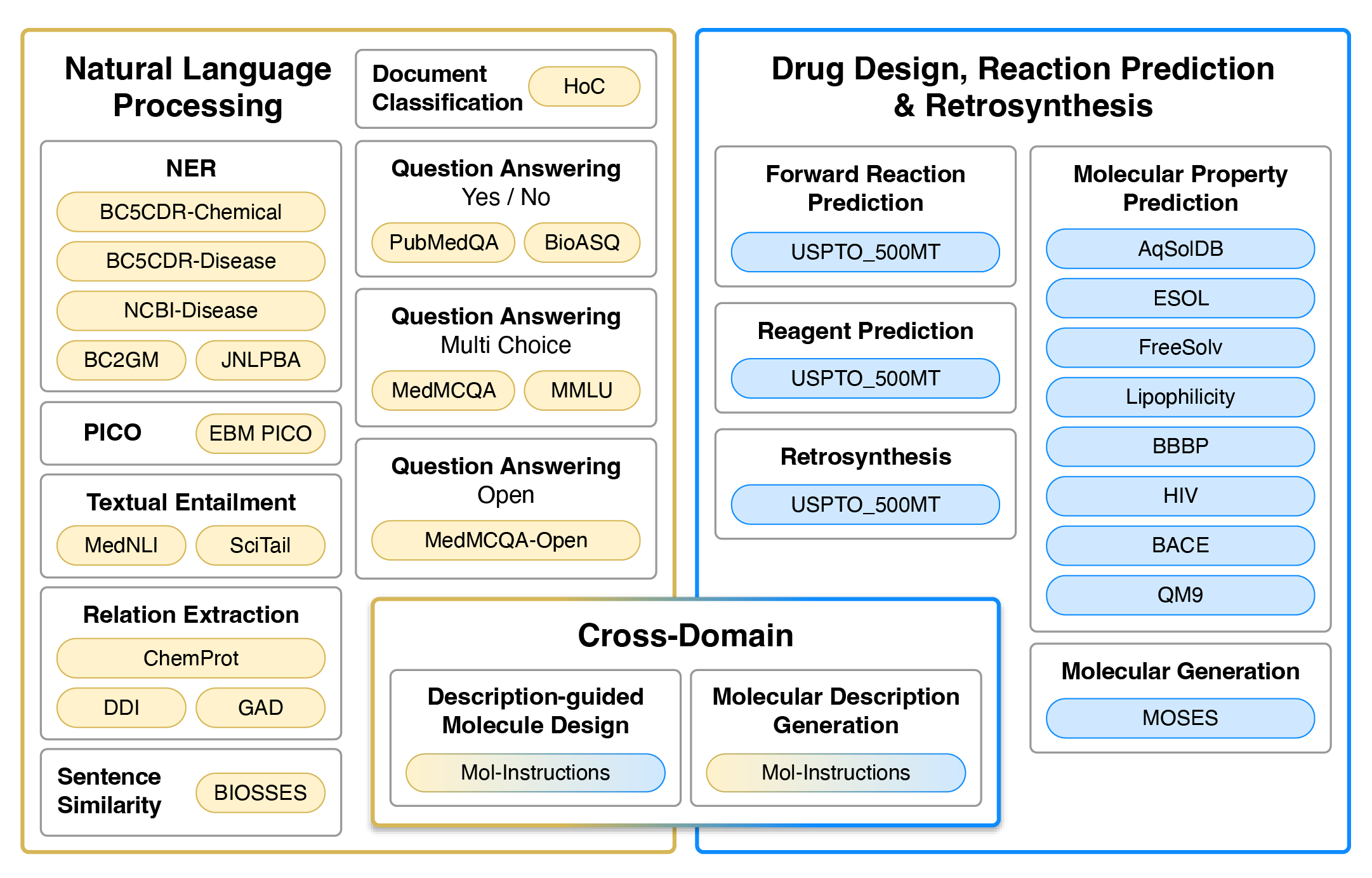
Tasks
Datasets used for training and evaluation. Colour represents the type of tasks. Yellow and blue datasets are single-domain, typically requiring regression/classification losses or generation in the target domain (natural language or SMILES strings). Gradients from yellow to blue represent cross-domain generation tasks that require natural language input and SMILES output, or vise versa.
Model Usage Guide
To use model for the inference follow the steps bellow:
1. Preprocess the input by replacing the atom tokens with special tokens.
```python
from transformers import AutoModelForSeq2SeqLM, AutoTokenizer
import re
from rdkit.Chem import MolFromSmiles
import string
from rdkit import RDLogger
RDLogger.DisableLog('rdApp.*')
atoms_tokens = ['Ag','Al','As','Au','B','Ba','Bi','Br','C','Ca',
'Cd','Cl','Co','Cr','Cs','Cu','F','Fe','Ga','Gd',
'Ge','H','Hg','I','In','K','Li','M','Mg','Mn',
'Mo','N','Na','O','P','Pt','Ru','S','Sb','Sc',
'Se','Si','Sn','V','W','Z','Zn','c','e','n','o','p','s']
atoms_tokens = sorted(atoms_tokens, key=lambda s: len(s), reverse=True)
SMI_REGEX_PATTERN = r"(\[|\]|\(|\)|\.|=|#|-|\+|\\|\/|:|~|@|\?|>>?|\*|\$|\%[0-9]{2}|[0-9]|" + \
'|'.join(atoms_tokens) + ")"
regex = re.compile(SMI_REGEX_PATTERN)
def clean_output_sequence(output_sequence):
return output_sequence.replace('', '').replace('', '').strip()
def add_special_symbols(text):
output = []
for word in text.split():
tokens = [token for token in regex.findall(word)]
if len(tokens) > 4 and (word == ''.join(tokens)) and MolFromSmiles(word):
output.append(''.join(['' for t in tokens]))
else:
output.append(word)
return ' '.join(output)
PROMPT = """Given the following reactants and reagents, please provide a possible product.
CCN(CC)CC.CCN=C=NCCCN(C)C.CN(C)C=O.Cl.NC1=CC=C(Cl)C=C1N.O.O=C(O)CCCCCNC(=O)C=C1C2=CC=CC=C2C2=CC=CC=C12.OC1=CC=CC2=C1N=NN2.[Cl-].[Na+]"""
PROMPT = add_special_symbols(PROMPT)
```
2. Load the model checkoint
```python
model = AutoModelForSeq2SeqLM.from_pretrained('insilicomedicine/nach0_base')
tokenizer = AutoTokenizer.from_pretrained('insilicomedicine/nach0_base')
```
3. Generate response to prompt and replace special tokens with corresponding atom tokens
```python
input_text_ids = tokenizer(PROMPT, padding="longest", max_length=512, truncation=True, return_tensors="pt")
generated_text_ids = model.generate(**input_text_ids, do_sample=True, top_k=100, top_p=0.95, max_length=512)
generated_text = tokenizer.batch_decode(generated_text_ids, skip_special_tokens=True)[0]
generated_text = clean_output_sequence(generated_text)
```
```python
# NC1=CC=C(Cl)C=C1NC(=O)CCCCCNC(=O)C=C1C2=CC=CC=C2C2=CC=CC=C12
```
References
If you use our repository, please cite the following related paper:
```
@inproceedings{....
}
```