---
license: apache-2.0
library_name: transformers
tags:
- genomics
- medical
datasets:
- kimou605/TATA-NOTATA-FineMistral-nucleotide_transformer_downstream_tasks
- InstaDeepAI/nucleotide_transformer_downstream_tasks
language:
- en
pipeline_tag: text-generation
---
 # ⚠️ STATE OF THE ART ⚠️
# Model Card for Model ID
BioTATA 7B V1 is a hybrid model merged between BioMistral 7B Dare and a 4bit QLORA adapter trained on TATA/NO TATA sequences from [InstaDeepAI nucleotide_transformer_downstream_tasks](https://huggingface.co/datasets/InstaDeepAI/nucleotide_transformer_downstream_tasks) dataset (promoters_all subset)
## Model Details
### Model Description
- **Developed by:** Karim Akkari (kimou605)
- **Model type:** FP32
- **Language(s) (NLP):** English
- **License:** Apache 2.0
- **Finetuned from model:** BioMistral 7B Dare
### Model Sources
- **Repository:** kimou605/BioTATA-7B
- **Demo:** [BioTATA 7B Space](https://huggingface.co/spaces/kimou605/BioTATA-7B)
## How to Get Started with the Model
```python
!pip install transformers
!pip install accelerate
!pip install bitsandbytes
```
```python
import os
import torch
import transformers
from transformers import (
AutoTokenizer,
AutoModelForCausalLM,
BitsAndBytesConfig,
pipeline
)
```
```python
model_name='kimou605/BioTATA-7B'
model_config = transformers.AutoConfig.from_pretrained(
model_name,
)
tokenizer = AutoTokenizer.from_pretrained(model_name, trust_remote_code=True)
tokenizer.pad_token = tokenizer.eos_token
tokenizer.padding_side = "right"
```
```python
# Activate 4-bit precision base model loading
use_4bit = True
# Compute dtype for 4-bit base models
bnb_4bit_compute_dtype = "float16"
# Quantization type (fp4 or nf4)
bnb_4bit_quant_type = "nf4"
# Activate nested quantization for 4-bit base models (double quantization)
use_nested_quant = True
```
```python
compute_dtype = getattr(torch, bnb_4bit_compute_dtype)
bnb_config = BitsAndBytesConfig(
load_in_4bit=use_4bit,
bnb_4bit_quant_type=bnb_4bit_quant_type,
bnb_4bit_compute_dtype=compute_dtype,
bnb_4bit_use_double_quant=use_nested_quant,
)
```
```python
model = AutoModelForCausalLM.from_pretrained(
model_name,
quantization_config=bnb_config,
)
```
```python
pipeline = transformers.pipeline(
"text-generation",
model=model,
torch_dtype=torch.float16,
device_map="auto",
tokenizer=tokenizer,
)
```
```python
messages = [{"role": "user", "content": "What is TATA"}]
prompt = tokenizer.apply_chat_template(messages, tokenize=False, add_generation_prompt=True)
outputs = pipeline(prompt, max_new_tokens=200, do_sample=True, temperature=0.01, top_k=50, top_p=0.95)
print(outputs[0]["generated_text"])
```
This will inference the model on 4.8GB Vram
## Bias, Risks, and Limitations
This model has been developped to show how can a medical LLM adapt itself to identify sequences as TATA/NO TATA
The adapter has been trained on a 53.3k rows for only 1 epoch (due to hardware limitations)
THIS MODEL IS FOR RESEARCH PURPOSES DO NOT USE IN PRODUCTION
### Recommendations
Users (both direct and downstream) should be made aware of the risks, biases and limitations of the model.
## Training Details
# ⚠️ STATE OF THE ART ⚠️
# Model Card for Model ID
BioTATA 7B V1 is a hybrid model merged between BioMistral 7B Dare and a 4bit QLORA adapter trained on TATA/NO TATA sequences from [InstaDeepAI nucleotide_transformer_downstream_tasks](https://huggingface.co/datasets/InstaDeepAI/nucleotide_transformer_downstream_tasks) dataset (promoters_all subset)
## Model Details
### Model Description
- **Developed by:** Karim Akkari (kimou605)
- **Model type:** FP32
- **Language(s) (NLP):** English
- **License:** Apache 2.0
- **Finetuned from model:** BioMistral 7B Dare
### Model Sources
- **Repository:** kimou605/BioTATA-7B
- **Demo:** [BioTATA 7B Space](https://huggingface.co/spaces/kimou605/BioTATA-7B)
## How to Get Started with the Model
```python
!pip install transformers
!pip install accelerate
!pip install bitsandbytes
```
```python
import os
import torch
import transformers
from transformers import (
AutoTokenizer,
AutoModelForCausalLM,
BitsAndBytesConfig,
pipeline
)
```
```python
model_name='kimou605/BioTATA-7B'
model_config = transformers.AutoConfig.from_pretrained(
model_name,
)
tokenizer = AutoTokenizer.from_pretrained(model_name, trust_remote_code=True)
tokenizer.pad_token = tokenizer.eos_token
tokenizer.padding_side = "right"
```
```python
# Activate 4-bit precision base model loading
use_4bit = True
# Compute dtype for 4-bit base models
bnb_4bit_compute_dtype = "float16"
# Quantization type (fp4 or nf4)
bnb_4bit_quant_type = "nf4"
# Activate nested quantization for 4-bit base models (double quantization)
use_nested_quant = True
```
```python
compute_dtype = getattr(torch, bnb_4bit_compute_dtype)
bnb_config = BitsAndBytesConfig(
load_in_4bit=use_4bit,
bnb_4bit_quant_type=bnb_4bit_quant_type,
bnb_4bit_compute_dtype=compute_dtype,
bnb_4bit_use_double_quant=use_nested_quant,
)
```
```python
model = AutoModelForCausalLM.from_pretrained(
model_name,
quantization_config=bnb_config,
)
```
```python
pipeline = transformers.pipeline(
"text-generation",
model=model,
torch_dtype=torch.float16,
device_map="auto",
tokenizer=tokenizer,
)
```
```python
messages = [{"role": "user", "content": "What is TATA"}]
prompt = tokenizer.apply_chat_template(messages, tokenize=False, add_generation_prompt=True)
outputs = pipeline(prompt, max_new_tokens=200, do_sample=True, temperature=0.01, top_k=50, top_p=0.95)
print(outputs[0]["generated_text"])
```
This will inference the model on 4.8GB Vram
## Bias, Risks, and Limitations
This model has been developped to show how can a medical LLM adapt itself to identify sequences as TATA/NO TATA
The adapter has been trained on a 53.3k rows for only 1 epoch (due to hardware limitations)
THIS MODEL IS FOR RESEARCH PURPOSES DO NOT USE IN PRODUCTION
### Recommendations
Users (both direct and downstream) should be made aware of the risks, biases and limitations of the model.
## Training Details
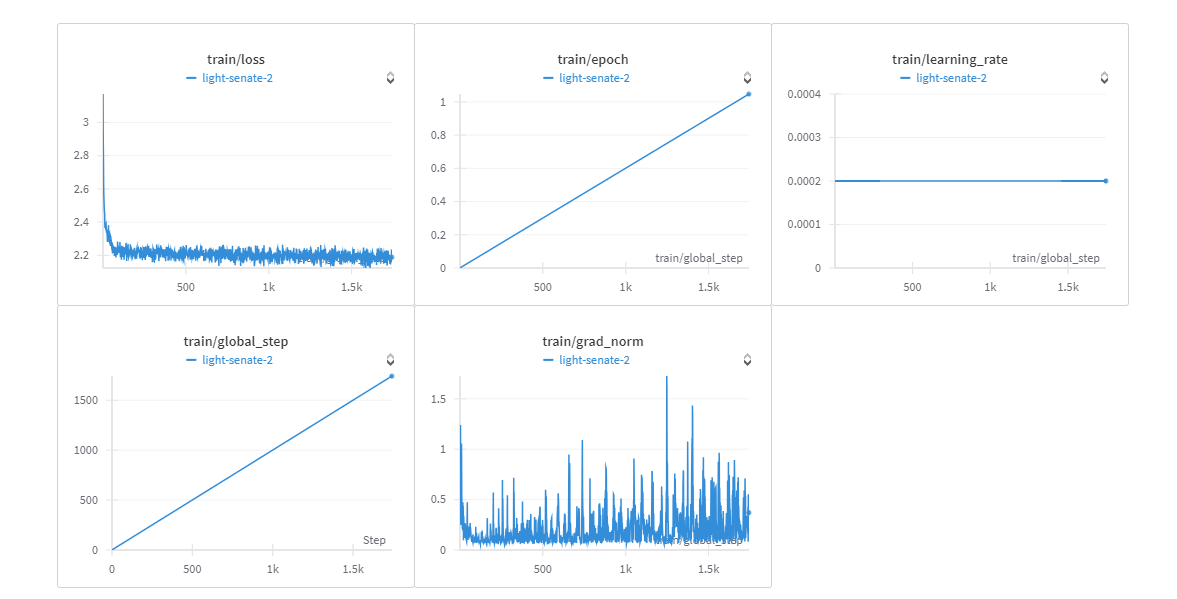
 You can view training report [here](https://wandb.ai/esprit-innovision/Fine%20tuning%20mistral%207B%20instadeep/reports/BioTATA--Vmlldzo3ODIwNTU3).
### Training Data
kimou605/TATA-NOTATA-FineMistral-nucleotide_transformer_downstream_tasks
### Training Procedure
#### Training Hyperparameters
- **Training regime:** BF16 4bits
#### Speeds, Sizes, Times
7h/ epoch
batch_per_gpu 32
GPU: NVIDIA A40 45GB Vram
## Environmental Impact
Carbon emissions can be estimated using the [Machine Learning Impact calculator](https://mlco2.github.io/impact#compute) presented in [Lacoste et al. (2019)](https://arxiv.org/abs/1910.09700).
- **Hardware Type:** NVIDIA A40
- **Hours used:** 11H
- **Cloud Provider:** vast.ai
- **Compute Region:** Europe
## Model Card Contact
Karim Akkari (kimou605)
You can view training report [here](https://wandb.ai/esprit-innovision/Fine%20tuning%20mistral%207B%20instadeep/reports/BioTATA--Vmlldzo3ODIwNTU3).
### Training Data
kimou605/TATA-NOTATA-FineMistral-nucleotide_transformer_downstream_tasks
### Training Procedure
#### Training Hyperparameters
- **Training regime:** BF16 4bits
#### Speeds, Sizes, Times
7h/ epoch
batch_per_gpu 32
GPU: NVIDIA A40 45GB Vram
## Environmental Impact
Carbon emissions can be estimated using the [Machine Learning Impact calculator](https://mlco2.github.io/impact#compute) presented in [Lacoste et al. (2019)](https://arxiv.org/abs/1910.09700).
- **Hardware Type:** NVIDIA A40
- **Hours used:** 11H
- **Cloud Provider:** vast.ai
- **Compute Region:** Europe
## Model Card Contact
Karim Akkari (kimou605) # ⚠️ STATE OF THE ART ⚠️
# Model Card for Model ID
BioTATA 7B V1 is a hybrid model merged between BioMistral 7B Dare and a 4bit QLORA adapter trained on TATA/NO TATA sequences from [InstaDeepAI nucleotide_transformer_downstream_tasks](https://huggingface.co/datasets/InstaDeepAI/nucleotide_transformer_downstream_tasks) dataset (promoters_all subset)
## Model Details
### Model Description
- **Developed by:** Karim Akkari (kimou605)
- **Model type:** FP32
- **Language(s) (NLP):** English
- **License:** Apache 2.0
- **Finetuned from model:** BioMistral 7B Dare
### Model Sources
- **Repository:** kimou605/BioTATA-7B
- **Demo:** [BioTATA 7B Space](https://huggingface.co/spaces/kimou605/BioTATA-7B)
## How to Get Started with the Model
```python
!pip install transformers
!pip install accelerate
!pip install bitsandbytes
```
```python
import os
import torch
import transformers
from transformers import (
AutoTokenizer,
AutoModelForCausalLM,
BitsAndBytesConfig,
pipeline
)
```
```python
model_name='kimou605/BioTATA-7B'
model_config = transformers.AutoConfig.from_pretrained(
model_name,
)
tokenizer = AutoTokenizer.from_pretrained(model_name, trust_remote_code=True)
tokenizer.pad_token = tokenizer.eos_token
tokenizer.padding_side = "right"
```
```python
# Activate 4-bit precision base model loading
use_4bit = True
# Compute dtype for 4-bit base models
bnb_4bit_compute_dtype = "float16"
# Quantization type (fp4 or nf4)
bnb_4bit_quant_type = "nf4"
# Activate nested quantization for 4-bit base models (double quantization)
use_nested_quant = True
```
```python
compute_dtype = getattr(torch, bnb_4bit_compute_dtype)
bnb_config = BitsAndBytesConfig(
load_in_4bit=use_4bit,
bnb_4bit_quant_type=bnb_4bit_quant_type,
bnb_4bit_compute_dtype=compute_dtype,
bnb_4bit_use_double_quant=use_nested_quant,
)
```
```python
model = AutoModelForCausalLM.from_pretrained(
model_name,
quantization_config=bnb_config,
)
```
```python
pipeline = transformers.pipeline(
"text-generation",
model=model,
torch_dtype=torch.float16,
device_map="auto",
tokenizer=tokenizer,
)
```
```python
messages = [{"role": "user", "content": "What is TATA"}]
prompt = tokenizer.apply_chat_template(messages, tokenize=False, add_generation_prompt=True)
outputs = pipeline(prompt, max_new_tokens=200, do_sample=True, temperature=0.01, top_k=50, top_p=0.95)
print(outputs[0]["generated_text"])
```
This will inference the model on 4.8GB Vram
## Bias, Risks, and Limitations
This model has been developped to show how can a medical LLM adapt itself to identify sequences as TATA/NO TATA
The adapter has been trained on a 53.3k rows for only 1 epoch (due to hardware limitations)
THIS MODEL IS FOR RESEARCH PURPOSES DO NOT USE IN PRODUCTION
### Recommendations
Users (both direct and downstream) should be made aware of the risks, biases and limitations of the model.
## Training Details
# ⚠️ STATE OF THE ART ⚠️
# Model Card for Model ID
BioTATA 7B V1 is a hybrid model merged between BioMistral 7B Dare and a 4bit QLORA adapter trained on TATA/NO TATA sequences from [InstaDeepAI nucleotide_transformer_downstream_tasks](https://huggingface.co/datasets/InstaDeepAI/nucleotide_transformer_downstream_tasks) dataset (promoters_all subset)
## Model Details
### Model Description
- **Developed by:** Karim Akkari (kimou605)
- **Model type:** FP32
- **Language(s) (NLP):** English
- **License:** Apache 2.0
- **Finetuned from model:** BioMistral 7B Dare
### Model Sources
- **Repository:** kimou605/BioTATA-7B
- **Demo:** [BioTATA 7B Space](https://huggingface.co/spaces/kimou605/BioTATA-7B)
## How to Get Started with the Model
```python
!pip install transformers
!pip install accelerate
!pip install bitsandbytes
```
```python
import os
import torch
import transformers
from transformers import (
AutoTokenizer,
AutoModelForCausalLM,
BitsAndBytesConfig,
pipeline
)
```
```python
model_name='kimou605/BioTATA-7B'
model_config = transformers.AutoConfig.from_pretrained(
model_name,
)
tokenizer = AutoTokenizer.from_pretrained(model_name, trust_remote_code=True)
tokenizer.pad_token = tokenizer.eos_token
tokenizer.padding_side = "right"
```
```python
# Activate 4-bit precision base model loading
use_4bit = True
# Compute dtype for 4-bit base models
bnb_4bit_compute_dtype = "float16"
# Quantization type (fp4 or nf4)
bnb_4bit_quant_type = "nf4"
# Activate nested quantization for 4-bit base models (double quantization)
use_nested_quant = True
```
```python
compute_dtype = getattr(torch, bnb_4bit_compute_dtype)
bnb_config = BitsAndBytesConfig(
load_in_4bit=use_4bit,
bnb_4bit_quant_type=bnb_4bit_quant_type,
bnb_4bit_compute_dtype=compute_dtype,
bnb_4bit_use_double_quant=use_nested_quant,
)
```
```python
model = AutoModelForCausalLM.from_pretrained(
model_name,
quantization_config=bnb_config,
)
```
```python
pipeline = transformers.pipeline(
"text-generation",
model=model,
torch_dtype=torch.float16,
device_map="auto",
tokenizer=tokenizer,
)
```
```python
messages = [{"role": "user", "content": "What is TATA"}]
prompt = tokenizer.apply_chat_template(messages, tokenize=False, add_generation_prompt=True)
outputs = pipeline(prompt, max_new_tokens=200, do_sample=True, temperature=0.01, top_k=50, top_p=0.95)
print(outputs[0]["generated_text"])
```
This will inference the model on 4.8GB Vram
## Bias, Risks, and Limitations
This model has been developped to show how can a medical LLM adapt itself to identify sequences as TATA/NO TATA
The adapter has been trained on a 53.3k rows for only 1 epoch (due to hardware limitations)
THIS MODEL IS FOR RESEARCH PURPOSES DO NOT USE IN PRODUCTION
### Recommendations
Users (both direct and downstream) should be made aware of the risks, biases and limitations of the model.
## Training Details

 You can view training report [here](https://wandb.ai/esprit-innovision/Fine%20tuning%20mistral%207B%20instadeep/reports/BioTATA--Vmlldzo3ODIwNTU3).
### Training Data
kimou605/TATA-NOTATA-FineMistral-nucleotide_transformer_downstream_tasks
### Training Procedure
#### Training Hyperparameters
- **Training regime:** BF16 4bits
#### Speeds, Sizes, Times
7h/ epoch
batch_per_gpu 32
GPU: NVIDIA A40 45GB Vram
## Environmental Impact
Carbon emissions can be estimated using the [Machine Learning Impact calculator](https://mlco2.github.io/impact#compute) presented in [Lacoste et al. (2019)](https://arxiv.org/abs/1910.09700).
- **Hardware Type:** NVIDIA A40
- **Hours used:** 11H
- **Cloud Provider:** vast.ai
- **Compute Region:** Europe
## Model Card Contact
Karim Akkari (kimou605)
You can view training report [here](https://wandb.ai/esprit-innovision/Fine%20tuning%20mistral%207B%20instadeep/reports/BioTATA--Vmlldzo3ODIwNTU3).
### Training Data
kimou605/TATA-NOTATA-FineMistral-nucleotide_transformer_downstream_tasks
### Training Procedure
#### Training Hyperparameters
- **Training regime:** BF16 4bits
#### Speeds, Sizes, Times
7h/ epoch
batch_per_gpu 32
GPU: NVIDIA A40 45GB Vram
## Environmental Impact
Carbon emissions can be estimated using the [Machine Learning Impact calculator](https://mlco2.github.io/impact#compute) presented in [Lacoste et al. (2019)](https://arxiv.org/abs/1910.09700).
- **Hardware Type:** NVIDIA A40
- **Hours used:** 11H
- **Cloud Provider:** vast.ai
- **Compute Region:** Europe
## Model Card Contact
Karim Akkari (kimou605)