restructure readme to match updated template
Browse files- README.md +44 -54
- configs/metadata.json +2 -1
- docs/README.md +44 -54
README.md
CHANGED
|
@@ -5,19 +5,26 @@ tags:
|
|
| 5 |
library_name: monai
|
| 6 |
license: apache-2.0
|
| 7 |
---
|
| 8 |
-
# Description
|
| 9 |
-
A neural architecture search algorithm for volumetric (3D) segmentation of the pancreas and pancreatic tumor from CT image.
|
| 10 |
-
|
| 11 |
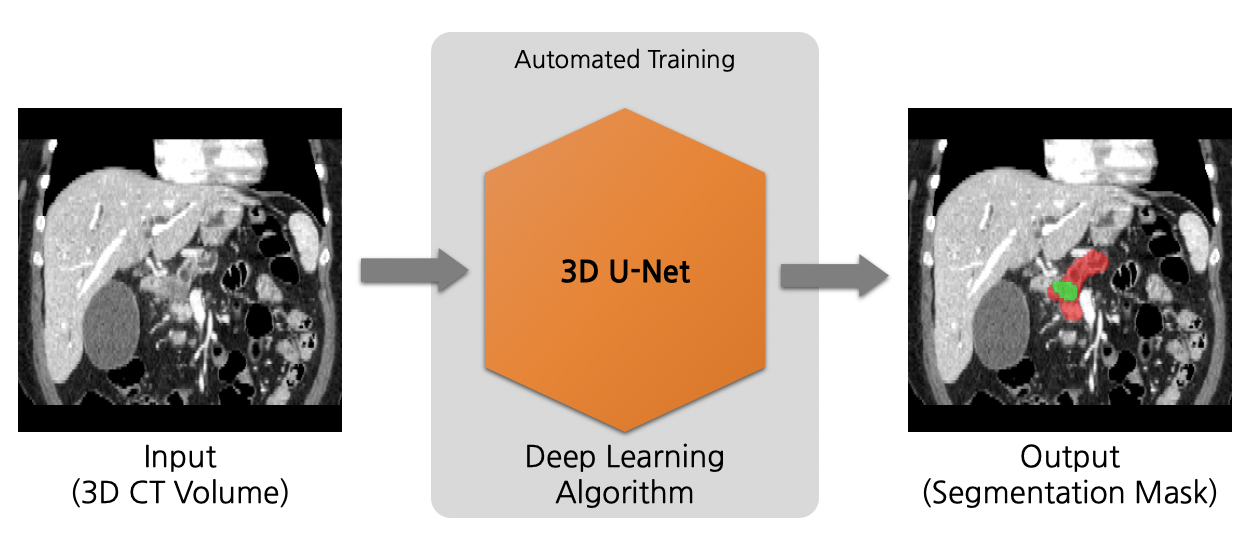
# Model Overview
|
| 12 |
-
|
| 13 |
-
|
| 14 |
-
This model is trained using the neural network model from the neural architecture search algorithm, DiNTS [1].
|
| 15 |
|
| 16 |
-

|
| 18 |
|
| 19 |
## Data
|
| 20 |
-
The training dataset is
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 21 |
|
| 22 |
## Training configuration
|
| 23 |
The training was performed with at least 16GB-memory GPUs.
|
|
@@ -32,7 +39,7 @@ The neural architecture search was performed with the following:
|
|
| 32 |
- Initial Learning Rate: 0.025
|
| 33 |
- Loss: DiceCELoss
|
| 34 |
|
| 35 |
-
### Training Configuration
|
| 36 |
The training was performed with the following:
|
| 37 |
|
| 38 |
- AMP: True
|
|
@@ -43,98 +50,81 @@ The training was performed with the following:
|
|
| 43 |
|
| 44 |
The segmentation of pancreas region is formulated as the voxel-wise 3-class classification. Each voxel is predicted as either foreground (pancreas body, tumour) or background. And the model is optimized with gradient descent method minimizing soft dice loss and cross-entropy loss between the predicted mask and ground truth segmentation.
|
| 45 |
|
| 46 |
-
###
|
| 47 |
-
|
| 48 |
-
|
| 49 |
|
| 50 |
-
|
| 51 |
-
|
| 52 |
-
-
|
| 53 |
-
-
|
| 54 |
-
-
|
| 55 |
-
- Randomly zooming volumes
|
| 56 |
-
- Randomly smoothing volumes with Gaussian kernels
|
| 57 |
-
- Randomly scaling intensity of the volume
|
| 58 |
-
- Randomly shifting intensity of the volume
|
| 59 |
-
- Randomly adding Gaussian noises
|
| 60 |
-
- Randomly flipping volumes
|
| 61 |
-
|
| 62 |
-
### Sliding-window Inference
|
| 63 |
-
Inference is performed in a sliding window manner with a specified stride.
|
| 64 |
-
|
| 65 |
-
## Input and output formats
|
| 66 |
-
Input: 1 channel CT image
|
| 67 |
-
|
| 68 |
-
Output: 3 channels: Label 2: pancreatic tumor; Label 1: pancreas; Label 0: everything else
|
| 69 |
|
| 70 |
## Performance
|
| 71 |
-
|
| 72 |
|
| 73 |
-
|
|
|
|
| 74 |
|
| 75 |
-
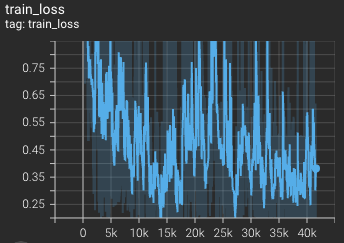
Training loss over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
|
| 76 |
|
| 77 |
-
|
|
|
|
| 78 |
|
| 79 |
-
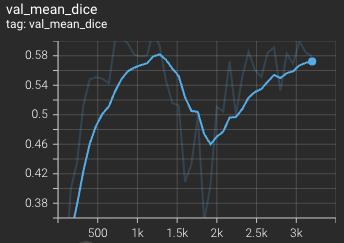
Validation mean dice score over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
|
| 80 |
-
|
| 81 |
-
|
| 82 |
|
| 83 |
### Searched Architecture Visualization
|
| 84 |
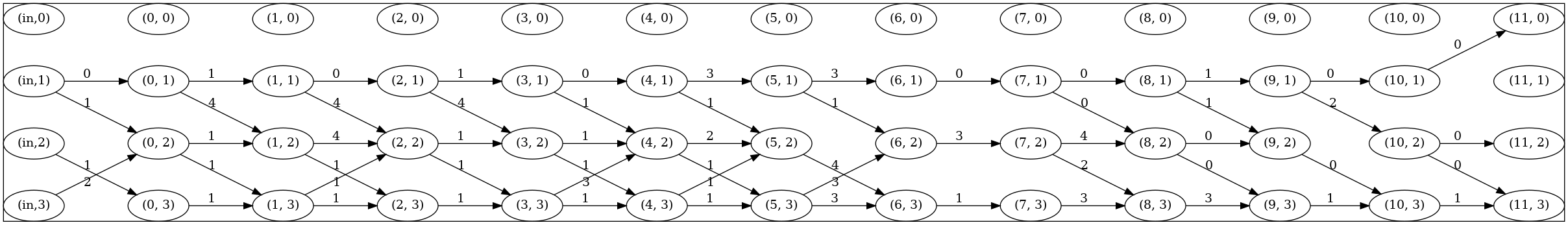
Users can install Graphviz for visualization of searched architectures (needed in custom/decode_plot.py). The edges between nodes indicate global structure, and numbers next to edges represent different operations in the cell searching space. An example of searched architecture is shown as follows:
|
| 85 |
|
| 86 |
-
|
| 87 |
-
Create data split (.json file):
|
| 88 |
|
| 89 |
-
|
| 90 |
-
|
| 91 |
-
```
|
| 92 |
|
| 93 |
-
|
|
|
|
|
|
|
| 94 |
|
| 95 |
```
|
| 96 |
python -m scripts.search run --config_file configs/search.yaml
|
| 97 |
```
|
| 98 |
|
| 99 |
-
Execute multi-GPU model searching (recommended):
|
| 100 |
|
| 101 |
```
|
| 102 |
torchrun --nnodes=1 --nproc_per_node=8 -m scripts.search run --config_file configs/search.yaml
|
| 103 |
```
|
| 104 |
|
| 105 |
-
Execute training:
|
| 106 |
|
| 107 |
```
|
| 108 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.yaml --logging_file configs/logging.conf
|
| 109 |
```
|
| 110 |
|
| 111 |
-
Override the `train` config to execute multi-GPU training:
|
| 112 |
|
| 113 |
```
|
| 114 |
torchrun --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.yaml','configs/multi_gpu_train.yaml']" --logging_file configs/logging.conf
|
| 115 |
```
|
| 116 |
|
| 117 |
-
Override the `train` config to execute evaluation with the trained model:
|
| 118 |
|
| 119 |
```
|
| 120 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.yaml','configs/evaluate.yaml']" --logging_file configs/logging.conf
|
| 121 |
```
|
| 122 |
|
| 123 |
-
Execute inference:
|
| 124 |
|
| 125 |
```
|
| 126 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.yaml --logging_file configs/logging.conf
|
| 127 |
```
|
| 128 |
|
| 129 |
-
Export checkpoint for TorchScript
|
| 130 |
|
| 131 |
```
|
| 132 |
python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.yaml
|
| 133 |
```
|
| 134 |
|
| 135 |
-
# Disclaimer
|
| 136 |
-
This is an example, not to be used for diagnostic purposes.
|
| 137 |
-
|
| 138 |
# References
|
| 139 |
[1] He, Y., Yang, D., Roth, H., Zhao, C. and Xu, D., 2021. Dints: Differentiable neural network topology search for 3d medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 5841-5850).
|
| 140 |
|
|
|
|
| 5 |
library_name: monai
|
| 6 |
license: apache-2.0
|
| 7 |
---
|
|
|
|
|
|
|
|
|
|
| 8 |
# Model Overview
|
| 9 |
+
A neural architecture search algorithm for volumetric (3D) segmentation of the pancreas and pancreatic tumor from CT image. This model is trained using the neural network model from the neural architecture search algorithm, DiNTS [1].
|
|
|
|
|
|
|
| 10 |
|
| 11 |
+
|
|
|
|
| 12 |
|
| 13 |
## Data
|
| 14 |
+
The training dataset is the Panceas Task from the Medical Segmentation Decathalon. Users can find more details on the datasets at http://medicaldecathlon.com/.
|
| 15 |
+
|
| 16 |
+
- Target: Liver and tumour
|
| 17 |
+
- Modality: Portal venous phase CT
|
| 18 |
+
- Size: 420 3D volumes (282 Training +139 Testing)
|
| 19 |
+
- Source: Memorial Sloan Kettering Cancer Center
|
| 20 |
+
- Challenge: Label unbalance with large (background), medium (pancreas) and small (tumour) structures.
|
| 21 |
+
|
| 22 |
+
### Preprocessing
|
| 23 |
+
The data list/split can be created with the script `scripts/prepare_datalist.py`.
|
| 24 |
+
|
| 25 |
+
```
|
| 26 |
+
python scripts/prepare_datalist.py --path /path-to-Task07_Pancreas/ --output configs/dataset_0.json
|
| 27 |
+
```
|
| 28 |
|
| 29 |
## Training configuration
|
| 30 |
The training was performed with at least 16GB-memory GPUs.
|
|
|
|
| 39 |
- Initial Learning Rate: 0.025
|
| 40 |
- Loss: DiceCELoss
|
| 41 |
|
| 42 |
+
### Optimial Architecture Training Configuration
|
| 43 |
The training was performed with the following:
|
| 44 |
|
| 45 |
- AMP: True
|
|
|
|
| 50 |
|
| 51 |
The segmentation of pancreas region is formulated as the voxel-wise 3-class classification. Each voxel is predicted as either foreground (pancreas body, tumour) or background. And the model is optimized with gradient descent method minimizing soft dice loss and cross-entropy loss between the predicted mask and ground truth segmentation.
|
| 52 |
|
| 53 |
+
### Input
|
| 54 |
+
One channel
|
| 55 |
+
- CT image
|
| 56 |
|
| 57 |
+
### Output
|
| 58 |
+
Three channels
|
| 59 |
+
- Label 2: pancreatic tumor
|
| 60 |
+
- Label 1: pancreas
|
| 61 |
+
- Label 0: everything else
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 62 |
|
| 63 |
## Performance
|
| 64 |
+
Dice score is used for evaluating the performance of the model. This model achieves a mean dice score of 0.62.
|
| 65 |
|
| 66 |
+
#### Training Loss
|
| 67 |
+
The loss over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
|
| 68 |
|
| 69 |
+
|
| 70 |
|
| 71 |
+
#### Validation Dice
|
| 72 |
+
The mean dice score over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
|
| 73 |
|
| 74 |
+

|
|
|
|
|
|
|
| 75 |
|
| 76 |
### Searched Architecture Visualization
|
| 77 |
Users can install Graphviz for visualization of searched architectures (needed in custom/decode_plot.py). The edges between nodes indicate global structure, and numbers next to edges represent different operations in the cell searching space. An example of searched architecture is shown as follows:
|
| 78 |
|
| 79 |
+
|
|
|
|
| 80 |
|
| 81 |
+
## MONAI Bundle Commands
|
| 82 |
+
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
|
|
|
|
| 83 |
|
| 84 |
+
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html).
|
| 85 |
+
|
| 86 |
+
#### Execute model searching:
|
| 87 |
|
| 88 |
```
|
| 89 |
python -m scripts.search run --config_file configs/search.yaml
|
| 90 |
```
|
| 91 |
|
| 92 |
+
#### Execute multi-GPU model searching (recommended):
|
| 93 |
|
| 94 |
```
|
| 95 |
torchrun --nnodes=1 --nproc_per_node=8 -m scripts.search run --config_file configs/search.yaml
|
| 96 |
```
|
| 97 |
|
| 98 |
+
#### Execute training:
|
| 99 |
|
| 100 |
```
|
| 101 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.yaml --logging_file configs/logging.conf
|
| 102 |
```
|
| 103 |
|
| 104 |
+
#### Override the `train` config to execute multi-GPU training:
|
| 105 |
|
| 106 |
```
|
| 107 |
torchrun --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.yaml','configs/multi_gpu_train.yaml']" --logging_file configs/logging.conf
|
| 108 |
```
|
| 109 |
|
| 110 |
+
#### Override the `train` config to execute evaluation with the trained model:
|
| 111 |
|
| 112 |
```
|
| 113 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.yaml','configs/evaluate.yaml']" --logging_file configs/logging.conf
|
| 114 |
```
|
| 115 |
|
| 116 |
+
#### Execute inference:
|
| 117 |
|
| 118 |
```
|
| 119 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.yaml --logging_file configs/logging.conf
|
| 120 |
```
|
| 121 |
|
| 122 |
+
#### Export checkpoint for TorchScript
|
| 123 |
|
| 124 |
```
|
| 125 |
python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.yaml
|
| 126 |
```
|
| 127 |
|
|
|
|
|
|
|
|
|
|
| 128 |
# References
|
| 129 |
[1] He, Y., Yang, D., Roth, H., Zhao, C. and Xu, D., 2021. Dints: Differentiable neural network topology search for 3d medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 5841-5850).
|
| 130 |
|
configs/metadata.json
CHANGED
|
@@ -1,7 +1,8 @@
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
-
"version": "0.3.
|
| 4 |
"changelog": {
|
|
|
|
| 5 |
"0.3.4": "correct typos",
|
| 6 |
"0.3.3": "update learning rate and readme",
|
| 7 |
"0.3.2": "update to use monai 1.0.1",
|
|
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.3.5",
|
| 4 |
"changelog": {
|
| 5 |
+
"0.3.5": "restructure readme to match updated template",
|
| 6 |
"0.3.4": "correct typos",
|
| 7 |
"0.3.3": "update learning rate and readme",
|
| 8 |
"0.3.2": "update to use monai 1.0.1",
|
docs/README.md
CHANGED
|
@@ -1,16 +1,23 @@
|
|
| 1 |
-
# Description
|
| 2 |
-
A neural architecture search algorithm for volumetric (3D) segmentation of the pancreas and pancreatic tumor from CT image.
|
| 3 |
-
|
| 4 |
# Model Overview
|
| 5 |
-
|
| 6 |
-
|
| 7 |
-
This model is trained using the neural network model from the neural architecture search algorithm, DiNTS [1].
|
| 8 |
|
| 9 |
-

|
| 11 |
|
| 12 |
## Data
|
| 13 |
-
The training dataset is
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 14 |
|
| 15 |
## Training configuration
|
| 16 |
The training was performed with at least 16GB-memory GPUs.
|
|
@@ -25,7 +32,7 @@ The neural architecture search was performed with the following:
|
|
| 25 |
- Initial Learning Rate: 0.025
|
| 26 |
- Loss: DiceCELoss
|
| 27 |
|
| 28 |
-
### Training Configuration
|
| 29 |
The training was performed with the following:
|
| 30 |
|
| 31 |
- AMP: True
|
|
@@ -36,98 +43,81 @@ The training was performed with the following:
|
|
| 36 |
|
| 37 |
The segmentation of pancreas region is formulated as the voxel-wise 3-class classification. Each voxel is predicted as either foreground (pancreas body, tumour) or background. And the model is optimized with gradient descent method minimizing soft dice loss and cross-entropy loss between the predicted mask and ground truth segmentation.
|
| 38 |
|
| 39 |
-
###
|
| 40 |
-
|
| 41 |
-
|
| 42 |
|
| 43 |
-
|
| 44 |
-
|
| 45 |
-
-
|
| 46 |
-
-
|
| 47 |
-
-
|
| 48 |
-
- Randomly zooming volumes
|
| 49 |
-
- Randomly smoothing volumes with Gaussian kernels
|
| 50 |
-
- Randomly scaling intensity of the volume
|
| 51 |
-
- Randomly shifting intensity of the volume
|
| 52 |
-
- Randomly adding Gaussian noises
|
| 53 |
-
- Randomly flipping volumes
|
| 54 |
-
|
| 55 |
-
### Sliding-window Inference
|
| 56 |
-
Inference is performed in a sliding window manner with a specified stride.
|
| 57 |
-
|
| 58 |
-
## Input and output formats
|
| 59 |
-
Input: 1 channel CT image
|
| 60 |
-
|
| 61 |
-
Output: 3 channels: Label 2: pancreatic tumor; Label 1: pancreas; Label 0: everything else
|
| 62 |
|
| 63 |
## Performance
|
| 64 |
-
|
| 65 |
|
| 66 |
-
|
|
|
|
| 67 |
|
| 68 |
-
Training loss over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
|
| 69 |
|
| 70 |
-
|
|
|
|
| 71 |
|
| 72 |
-
Validation mean dice score over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
|
| 73 |
-
|
| 74 |
-

|
| 75 |
|
| 76 |
### Searched Architecture Visualization
|
| 77 |
Users can install Graphviz for visualization of searched architectures (needed in custom/decode_plot.py). The edges between nodes indicate global structure, and numbers next to edges represent different operations in the cell searching space. An example of searched architecture is shown as follows:
|
| 78 |
|
| 79 |
-
|
| 80 |
-
Create data split (.json file):
|
| 81 |
|
| 82 |
-
|
| 83 |
-
|
| 84 |
-
```
|
| 85 |
|
| 86 |
-
|
|
|
|
|
|
|
| 87 |
|
| 88 |
```
|
| 89 |
python -m scripts.search run --config_file configs/search.yaml
|
| 90 |
```
|
| 91 |
|
| 92 |
-
Execute multi-GPU model searching (recommended):
|
| 93 |
|
| 94 |
```
|
| 95 |
torchrun --nnodes=1 --nproc_per_node=8 -m scripts.search run --config_file configs/search.yaml
|
| 96 |
```
|
| 97 |
|
| 98 |
-
Execute training:
|
| 99 |
|
| 100 |
```
|
| 101 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.yaml --logging_file configs/logging.conf
|
| 102 |
```
|
| 103 |
|
| 104 |
-
Override the `train` config to execute multi-GPU training:
|
| 105 |
|
| 106 |
```
|
| 107 |
torchrun --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.yaml','configs/multi_gpu_train.yaml']" --logging_file configs/logging.conf
|
| 108 |
```
|
| 109 |
|
| 110 |
-
Override the `train` config to execute evaluation with the trained model:
|
| 111 |
|
| 112 |
```
|
| 113 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.yaml','configs/evaluate.yaml']" --logging_file configs/logging.conf
|
| 114 |
```
|
| 115 |
|
| 116 |
-
Execute inference:
|
| 117 |
|
| 118 |
```
|
| 119 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.yaml --logging_file configs/logging.conf
|
| 120 |
```
|
| 121 |
|
| 122 |
-
Export checkpoint for TorchScript
|
| 123 |
|
| 124 |
```
|
| 125 |
python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.yaml
|
| 126 |
```
|
| 127 |
|
| 128 |
-
# Disclaimer
|
| 129 |
-
This is an example, not to be used for diagnostic purposes.
|
| 130 |
-
|
| 131 |
# References
|
| 132 |
[1] He, Y., Yang, D., Roth, H., Zhao, C. and Xu, D., 2021. Dints: Differentiable neural network topology search for 3d medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 5841-5850).
|
| 133 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
# Model Overview
|
| 2 |
+
A neural architecture search algorithm for volumetric (3D) segmentation of the pancreas and pancreatic tumor from CT image. This model is trained using the neural network model from the neural architecture search algorithm, DiNTS [1].
|
|
|
|
|
|
|
| 3 |
|
| 4 |
+

|
|
|
|
| 5 |
|
| 6 |
## Data
|
| 7 |
+
The training dataset is the Panceas Task from the Medical Segmentation Decathalon. Users can find more details on the datasets at http://medicaldecathlon.com/.
|
| 8 |
+
|
| 9 |
+
- Target: Liver and tumour
|
| 10 |
+
- Modality: Portal venous phase CT
|
| 11 |
+
- Size: 420 3D volumes (282 Training +139 Testing)
|
| 12 |
+
- Source: Memorial Sloan Kettering Cancer Center
|
| 13 |
+
- Challenge: Label unbalance with large (background), medium (pancreas) and small (tumour) structures.
|
| 14 |
+
|
| 15 |
+
### Preprocessing
|
| 16 |
+
The data list/split can be created with the script `scripts/prepare_datalist.py`.
|
| 17 |
+
|
| 18 |
+
```
|
| 19 |
+
python scripts/prepare_datalist.py --path /path-to-Task07_Pancreas/ --output configs/dataset_0.json
|
| 20 |
+
```
|
| 21 |
|
| 22 |
## Training configuration
|
| 23 |
The training was performed with at least 16GB-memory GPUs.
|
|
|
|
| 32 |
- Initial Learning Rate: 0.025
|
| 33 |
- Loss: DiceCELoss
|
| 34 |
|
| 35 |
+
### Optimial Architecture Training Configuration
|
| 36 |
The training was performed with the following:
|
| 37 |
|
| 38 |
- AMP: True
|
|
|
|
| 43 |
|
| 44 |
The segmentation of pancreas region is formulated as the voxel-wise 3-class classification. Each voxel is predicted as either foreground (pancreas body, tumour) or background. And the model is optimized with gradient descent method minimizing soft dice loss and cross-entropy loss between the predicted mask and ground truth segmentation.
|
| 45 |
|
| 46 |
+
### Input
|
| 47 |
+
One channel
|
| 48 |
+
- CT image
|
| 49 |
|
| 50 |
+
### Output
|
| 51 |
+
Three channels
|
| 52 |
+
- Label 2: pancreatic tumor
|
| 53 |
+
- Label 1: pancreas
|
| 54 |
+
- Label 0: everything else
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 55 |
|
| 56 |
## Performance
|
| 57 |
+
Dice score is used for evaluating the performance of the model. This model achieves a mean dice score of 0.62.
|
| 58 |
|
| 59 |
+
#### Training Loss
|
| 60 |
+
The loss over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
|
| 61 |
|
| 62 |
+

|
| 63 |
|
| 64 |
+
#### Validation Dice
|
| 65 |
+
The mean dice score over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
|
| 66 |
|
| 67 |
+

|
|
|
|
|
|
|
| 68 |
|
| 69 |
### Searched Architecture Visualization
|
| 70 |
Users can install Graphviz for visualization of searched architectures (needed in custom/decode_plot.py). The edges between nodes indicate global structure, and numbers next to edges represent different operations in the cell searching space. An example of searched architecture is shown as follows:
|
| 71 |
|
| 72 |
+

|
|
|
|
| 73 |
|
| 74 |
+
## MONAI Bundle Commands
|
| 75 |
+
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
|
|
|
|
| 76 |
|
| 77 |
+
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html).
|
| 78 |
+
|
| 79 |
+
#### Execute model searching:
|
| 80 |
|
| 81 |
```
|
| 82 |
python -m scripts.search run --config_file configs/search.yaml
|
| 83 |
```
|
| 84 |
|
| 85 |
+
#### Execute multi-GPU model searching (recommended):
|
| 86 |
|
| 87 |
```
|
| 88 |
torchrun --nnodes=1 --nproc_per_node=8 -m scripts.search run --config_file configs/search.yaml
|
| 89 |
```
|
| 90 |
|
| 91 |
+
#### Execute training:
|
| 92 |
|
| 93 |
```
|
| 94 |
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.yaml --logging_file configs/logging.conf
|
| 95 |
```
|
| 96 |
|
| 97 |
+
#### Override the `train` config to execute multi-GPU training:
|
| 98 |
|
| 99 |
```
|
| 100 |
torchrun --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.yaml','configs/multi_gpu_train.yaml']" --logging_file configs/logging.conf
|
| 101 |
```
|
| 102 |
|
| 103 |
+
#### Override the `train` config to execute evaluation with the trained model:
|
| 104 |
|
| 105 |
```
|
| 106 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.yaml','configs/evaluate.yaml']" --logging_file configs/logging.conf
|
| 107 |
```
|
| 108 |
|
| 109 |
+
#### Execute inference:
|
| 110 |
|
| 111 |
```
|
| 112 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.yaml --logging_file configs/logging.conf
|
| 113 |
```
|
| 114 |
|
| 115 |
+
#### Export checkpoint for TorchScript
|
| 116 |
|
| 117 |
```
|
| 118 |
python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.yaml
|
| 119 |
```
|
| 120 |
|
|
|
|
|
|
|
|
|
|
| 121 |
# References
|
| 122 |
[1] He, Y., Yang, D., Roth, H., Zhao, C. and Xu, D., 2021. Dints: Differentiable neural network topology search for 3d medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 5841-5850).
|
| 123 |
|