update learning rate and readme
Browse files- README.md +63 -3
- configs/metadata.json +2 -1
- configs/multi_gpu_train.yaml +1 -1
- docs/README.md +63 -3
- models/model.pt +2 -2
- models/model.ts +2 -2
- models/search_code_18590.pt +2 -2
README.md
CHANGED
|
@@ -11,6 +11,11 @@ A neural architecture search algorithm for volumetric (3D) segmentation of the p
|
|
| 11 |
# Model Overview
|
| 12 |
This model is trained using the state-of-the-art algorithm [1] of the "Medical Segmentation Decathlon Challenge 2018" with 196 training images, 56 validation images, and 28 testing images.
|
| 13 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 14 |
## Data
|
| 15 |
The training dataset is Task07_Pancreas.tar from http://medicaldecathlon.com/. And the data list/split can be created with the script `scripts/prepare_datalist.py`.
|
| 16 |
|
|
@@ -19,17 +24,66 @@ The training was performed with at least 16GB-memory GPUs.
|
|
| 19 |
|
| 20 |
Actual Model Input: 96 x 96 x 96
|
| 21 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 22 |
## Input and output formats
|
| 23 |
Input: 1 channel CT image
|
| 24 |
|
| 25 |
Output: 3 channels: Label 2: pancreatic tumor; Label 1: pancreas; Label 0: everything else
|
| 26 |
|
| 27 |
-
##
|
| 28 |
This model achieves the following Dice score on the validation data (our own split from the training dataset):
|
| 29 |
|
| 30 |
-
Mean Dice = 0.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 31 |
|
| 32 |
-
|
|
|
|
|
|
|
|
|
|
| 33 |
Create data split (.json file):
|
| 34 |
|
| 35 |
```
|
|
@@ -72,6 +126,12 @@ Execute inference:
|
|
| 72 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.yaml --logging_file configs/logging.conf
|
| 73 |
```
|
| 74 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 75 |
# Disclaimer
|
| 76 |
This is an example, not to be used for diagnostic purposes.
|
| 77 |
|
|
|
|
| 11 |
# Model Overview
|
| 12 |
This model is trained using the state-of-the-art algorithm [1] of the "Medical Segmentation Decathlon Challenge 2018" with 196 training images, 56 validation images, and 28 testing images.
|
| 13 |
|
| 14 |
+
This model is trained using the neural network model from the neural architecture search algorithm, DiNTS [1].
|
| 15 |
+
|
| 16 |
+

|
| 18 |
+
|
| 19 |
## Data
|
| 20 |
The training dataset is Task07_Pancreas.tar from http://medicaldecathlon.com/. And the data list/split can be created with the script `scripts/prepare_datalist.py`.
|
| 21 |
|
|
|
|
| 24 |
|
| 25 |
Actual Model Input: 96 x 96 x 96
|
| 26 |
|
| 27 |
+
### Neural Architecture Search Configuration
|
| 28 |
+
The neural architecture search was performed with the following:
|
| 29 |
+
|
| 30 |
+
- AMP: True
|
| 31 |
+
- Optimizer: SGD
|
| 32 |
+
- Initial Learning Rate: 0.025
|
| 33 |
+
- Loss: DiceCELoss
|
| 34 |
+
|
| 35 |
+
### Training Configuration
|
| 36 |
+
The training was performed with the following:
|
| 37 |
+
|
| 38 |
+
- AMP: True
|
| 39 |
+
- Optimizer: SGD
|
| 40 |
+
- (Initial) Learning Rate: 0.025
|
| 41 |
+
- Loss: DiceCELoss
|
| 42 |
+
- Note: If out-of-memory or program crash occurs while caching the data set, please change the cache\_rate in CacheDataset to a lower value in the range (0, 1).
|
| 43 |
+
|
| 44 |
+
The segmentation of pancreas region is formulated as the voxel-wise 3-class classification. Each voxel is predicted as either foreground (pancreas body, tumour) or background. And the model is optimized with gradient descent method minimizing soft dice loss and cross-entropy loss between the predicted mask and ground truth segmentation.
|
| 45 |
+
|
| 46 |
+
### Data Pre-processing and Augmentation
|
| 47 |
+
|
| 48 |
+
Input: 1 channel CT image with intensity in HU
|
| 49 |
+
|
| 50 |
+
- Converting to channel first
|
| 51 |
+
- Normalizing and clipping intensities of tissue window to [0,1]
|
| 52 |
+
- Cropping foreground surrounding regions
|
| 53 |
+
- Cropping random fixed sized regions of size [96, 96, 96] with the center being a foreground or background voxel at ratio 1 : 1
|
| 54 |
+
- Randomly rotating volumes
|
| 55 |
+
- Randomly zooming volumes
|
| 56 |
+
- Randomly smoothing volumes with Gaussian kernels
|
| 57 |
+
- Randomly scaling intensity of the volume
|
| 58 |
+
- Randomly shifting intensity of the volume
|
| 59 |
+
- Randomly adding Gaussian noises
|
| 60 |
+
- Randomly flipping volumes
|
| 61 |
+
|
| 62 |
+
### Sliding-window Inference
|
| 63 |
+
Inference is performed in a sliding window manner with a specified stride.
|
| 64 |
+
|
| 65 |
## Input and output formats
|
| 66 |
Input: 1 channel CT image
|
| 67 |
|
| 68 |
Output: 3 channels: Label 2: pancreatic tumor; Label 1: pancreas; Label 0: everything else
|
| 69 |
|
| 70 |
+
## Performance
|
| 71 |
This model achieves the following Dice score on the validation data (our own split from the training dataset):
|
| 72 |
|
| 73 |
+
Mean Dice = 0.62
|
| 74 |
+
|
| 75 |
+
Training loss over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
|
| 76 |
+
|
| 77 |
+
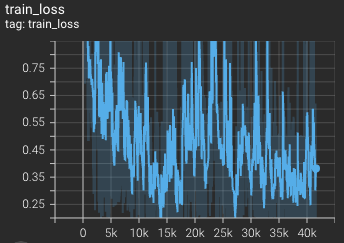
|
| 78 |
+
|
| 79 |
+
Validation mean dice score over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
|
| 80 |
+
|
| 81 |
+
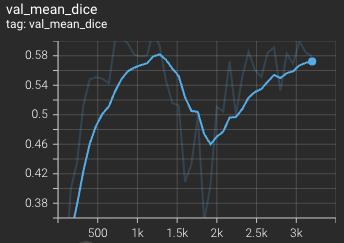
|
| 82 |
|
| 83 |
+
### Searched Architecture Visualization
|
| 84 |
+
Users can install Graphviz for visualization of searched architectures (needed in custom/decode_plot.py). The edges between nodes indicate global structure, and numbers next to edges represent different operations in the cell searching space. An example of searched architecture is shown as follows:
|
| 85 |
+
|
| 86 |
+
## Commands Example
|
| 87 |
Create data split (.json file):
|
| 88 |
|
| 89 |
```
|
|
|
|
| 126 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.yaml --logging_file configs/logging.conf
|
| 127 |
```
|
| 128 |
|
| 129 |
+
Export checkpoint for TorchScript
|
| 130 |
+
|
| 131 |
+
```
|
| 132 |
+
python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.yaml
|
| 133 |
+
```
|
| 134 |
+
|
| 135 |
# Disclaimer
|
| 136 |
This is an example, not to be used for diagnostic purposes.
|
| 137 |
|
configs/metadata.json
CHANGED
|
@@ -1,7 +1,8 @@
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
-
"version": "0.3.
|
| 4 |
"changelog": {
|
|
|
|
| 5 |
"0.3.2": "update to use monai 1.0.1",
|
| 6 |
"0.3.1": "fix license Copyright error",
|
| 7 |
"0.3.0": "update license files",
|
|
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.3.3",
|
| 4 |
"changelog": {
|
| 5 |
+
"0.3.3": "update learning rate and readme",
|
| 6 |
"0.3.2": "update to use monai 1.0.1",
|
| 7 |
"0.3.1": "fix license Copyright error",
|
| 8 |
"0.3.0": "update license files",
|
configs/multi_gpu_train.yaml
CHANGED
|
@@ -6,7 +6,7 @@ network:
|
|
| 6 |
find_unused_parameters: true
|
| 7 |
device_ids:
|
| 8 |
- "@device"
|
| 9 |
-
optimizer#lr: "$0.
|
| 10 |
lr_scheduler#step_size: "$80*dist.get_world_size()"
|
| 11 |
train#handlers:
|
| 12 |
- _target_: LrScheduleHandler
|
|
|
|
| 6 |
find_unused_parameters: true
|
| 7 |
device_ids:
|
| 8 |
- "@device"
|
| 9 |
+
optimizer#lr: "$0.025*dist.get_world_size()"
|
| 10 |
lr_scheduler#step_size: "$80*dist.get_world_size()"
|
| 11 |
train#handlers:
|
| 12 |
- _target_: LrScheduleHandler
|
docs/README.md
CHANGED
|
@@ -4,6 +4,11 @@ A neural architecture search algorithm for volumetric (3D) segmentation of the p
|
|
| 4 |
# Model Overview
|
| 5 |
This model is trained using the state-of-the-art algorithm [1] of the "Medical Segmentation Decathlon Challenge 2018" with 196 training images, 56 validation images, and 28 testing images.
|
| 6 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 7 |
## Data
|
| 8 |
The training dataset is Task07_Pancreas.tar from http://medicaldecathlon.com/. And the data list/split can be created with the script `scripts/prepare_datalist.py`.
|
| 9 |
|
|
@@ -12,17 +17,66 @@ The training was performed with at least 16GB-memory GPUs.
|
|
| 12 |
|
| 13 |
Actual Model Input: 96 x 96 x 96
|
| 14 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 15 |
## Input and output formats
|
| 16 |
Input: 1 channel CT image
|
| 17 |
|
| 18 |
Output: 3 channels: Label 2: pancreatic tumor; Label 1: pancreas; Label 0: everything else
|
| 19 |
|
| 20 |
-
##
|
| 21 |
This model achieves the following Dice score on the validation data (our own split from the training dataset):
|
| 22 |
|
| 23 |
-
Mean Dice = 0.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 24 |
|
| 25 |
-
|
|
|
|
|
|
|
|
|
|
| 26 |
Create data split (.json file):
|
| 27 |
|
| 28 |
```
|
|
@@ -65,6 +119,12 @@ Execute inference:
|
|
| 65 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.yaml --logging_file configs/logging.conf
|
| 66 |
```
|
| 67 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 68 |
# Disclaimer
|
| 69 |
This is an example, not to be used for diagnostic purposes.
|
| 70 |
|
|
|
|
| 4 |
# Model Overview
|
| 5 |
This model is trained using the state-of-the-art algorithm [1] of the "Medical Segmentation Decathlon Challenge 2018" with 196 training images, 56 validation images, and 28 testing images.
|
| 6 |
|
| 7 |
+
This model is trained using the neural network model from the neural architecture search algorithm, DiNTS [1].
|
| 8 |
+
|
| 9 |
+

|
| 11 |
+
|
| 12 |
## Data
|
| 13 |
The training dataset is Task07_Pancreas.tar from http://medicaldecathlon.com/. And the data list/split can be created with the script `scripts/prepare_datalist.py`.
|
| 14 |
|
|
|
|
| 17 |
|
| 18 |
Actual Model Input: 96 x 96 x 96
|
| 19 |
|
| 20 |
+
### Neural Architecture Search Configuration
|
| 21 |
+
The neural architecture search was performed with the following:
|
| 22 |
+
|
| 23 |
+
- AMP: True
|
| 24 |
+
- Optimizer: SGD
|
| 25 |
+
- Initial Learning Rate: 0.025
|
| 26 |
+
- Loss: DiceCELoss
|
| 27 |
+
|
| 28 |
+
### Training Configuration
|
| 29 |
+
The training was performed with the following:
|
| 30 |
+
|
| 31 |
+
- AMP: True
|
| 32 |
+
- Optimizer: SGD
|
| 33 |
+
- (Initial) Learning Rate: 0.025
|
| 34 |
+
- Loss: DiceCELoss
|
| 35 |
+
- Note: If out-of-memory or program crash occurs while caching the data set, please change the cache\_rate in CacheDataset to a lower value in the range (0, 1).
|
| 36 |
+
|
| 37 |
+
The segmentation of pancreas region is formulated as the voxel-wise 3-class classification. Each voxel is predicted as either foreground (pancreas body, tumour) or background. And the model is optimized with gradient descent method minimizing soft dice loss and cross-entropy loss between the predicted mask and ground truth segmentation.
|
| 38 |
+
|
| 39 |
+
### Data Pre-processing and Augmentation
|
| 40 |
+
|
| 41 |
+
Input: 1 channel CT image with intensity in HU
|
| 42 |
+
|
| 43 |
+
- Converting to channel first
|
| 44 |
+
- Normalizing and clipping intensities of tissue window to [0,1]
|
| 45 |
+
- Cropping foreground surrounding regions
|
| 46 |
+
- Cropping random fixed sized regions of size [96, 96, 96] with the center being a foreground or background voxel at ratio 1 : 1
|
| 47 |
+
- Randomly rotating volumes
|
| 48 |
+
- Randomly zooming volumes
|
| 49 |
+
- Randomly smoothing volumes with Gaussian kernels
|
| 50 |
+
- Randomly scaling intensity of the volume
|
| 51 |
+
- Randomly shifting intensity of the volume
|
| 52 |
+
- Randomly adding Gaussian noises
|
| 53 |
+
- Randomly flipping volumes
|
| 54 |
+
|
| 55 |
+
### Sliding-window Inference
|
| 56 |
+
Inference is performed in a sliding window manner with a specified stride.
|
| 57 |
+
|
| 58 |
## Input and output formats
|
| 59 |
Input: 1 channel CT image
|
| 60 |
|
| 61 |
Output: 3 channels: Label 2: pancreatic tumor; Label 1: pancreas; Label 0: everything else
|
| 62 |
|
| 63 |
+
## Performance
|
| 64 |
This model achieves the following Dice score on the validation data (our own split from the training dataset):
|
| 65 |
|
| 66 |
+
Mean Dice = 0.62
|
| 67 |
+
|
| 68 |
+
Training loss over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
|
| 69 |
+
|
| 70 |
+

|
| 71 |
+
|
| 72 |
+
Validation mean dice score over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
|
| 73 |
+
|
| 74 |
+

|
| 75 |
|
| 76 |
+
### Searched Architecture Visualization
|
| 77 |
+
Users can install Graphviz for visualization of searched architectures (needed in custom/decode_plot.py). The edges between nodes indicate global structure, and numbers next to edges represent different operations in the cell searching space. An example of searched architecture is shown as follows:
|
| 78 |
+
|
| 79 |
+
## Commands Example
|
| 80 |
Create data split (.json file):
|
| 81 |
|
| 82 |
```
|
|
|
|
| 119 |
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.yaml --logging_file configs/logging.conf
|
| 120 |
```
|
| 121 |
|
| 122 |
+
Export checkpoint for TorchScript
|
| 123 |
+
|
| 124 |
+
```
|
| 125 |
+
python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.yaml
|
| 126 |
+
```
|
| 127 |
+
|
| 128 |
# Disclaimer
|
| 129 |
This is an example, not to be used for diagnostic purposes.
|
| 130 |
|
models/model.pt
CHANGED
|
@@ -1,3 +1,3 @@
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
-
oid sha256:
|
| 3 |
-
size
|
|
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:4de79b954bb197c9a75d198c7b1038ad504cf0e370ae8b055c49c134c7b0e883
|
| 3 |
+
size 534788757
|
models/model.ts
CHANGED
|
@@ -1,3 +1,3 @@
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
-
oid sha256:
|
| 3 |
-
size
|
|
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:d9c47d9b4d1ec457dfa28c9e03c4ab847cdbe6ca825e3a84a245ca8a9caa631f
|
| 3 |
+
size 534978281
|
models/search_code_18590.pt
CHANGED
|
@@ -1,3 +1,3 @@
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
-
oid sha256:
|
| 3 |
-
size
|
|
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:21a2a05b173e9a5a80a009ce4baa3dfd8118463e7c26a1ef49cce573a3e662cc
|
| 3 |
+
size 4355
|