---
license: bsd-3-clause
tags:
- Confocal Fluorescence Microscopy
- Image Super-resolution
- Deep Learning
- Benchmark
---
# [SR-CACO-2: A Dataset for Confocal Fluorescence Microscopy Image Super-Resolution (NeurIPS2024)](https://arxiv.org/pdf/2406.09168.pdf)
by **Soufiane Belharbi1, Mara KM Whitford2,3,
Phuong Hoang2, Shakeeb Murtaza1, Luke McCaffrey2,3,4, Eric Granger1**
1 LIVIA, Dept. of Systems Engineering, ETS Montreal, Canada
2 Goodman Cancer Institute, McGill University, Montreal, Canada
3 Dept. of Biochemistry, McGill University, Montreal, Canada
4 Gerald Bronfman Dept. of Oncology, McGill University, Montreal,
Canada
[](https://arxiv.org/pdf/2406.09168)
[](https://github.com/sbelharbi/sr-caco-2)
## Abstract
Confocal fluorescence microscopy is one of the most accessible and widely used
imaging techniques for the study of biological processes at the cellular and
subcellular levels. Scanning confocal microscopy allows the capture of
high-quality images from thick three-dimensional (3D) samples, yet suffers from
well-known limitations such as photobleaching and phototoxicity of specimens
caused by intense light exposure, which limits its use in some applications,
especially for living cells. Cellular damage can be alleviated by changing
imaging parameters to reduce light exposure, often at the expense of image
quality. Machine/deep learning methods for single-image super-resolution (SISR)
can be applied to restore image quality by upscaling lower-resolution (LR)
images to produce high-resolution images (HR). These SISR methods have been
successfully applied to photo-realistic images due partly to the abundance of
publicly available data. In contrast, the lack of publicly available data
partly limits their application and success in scanning confocal microscopy.
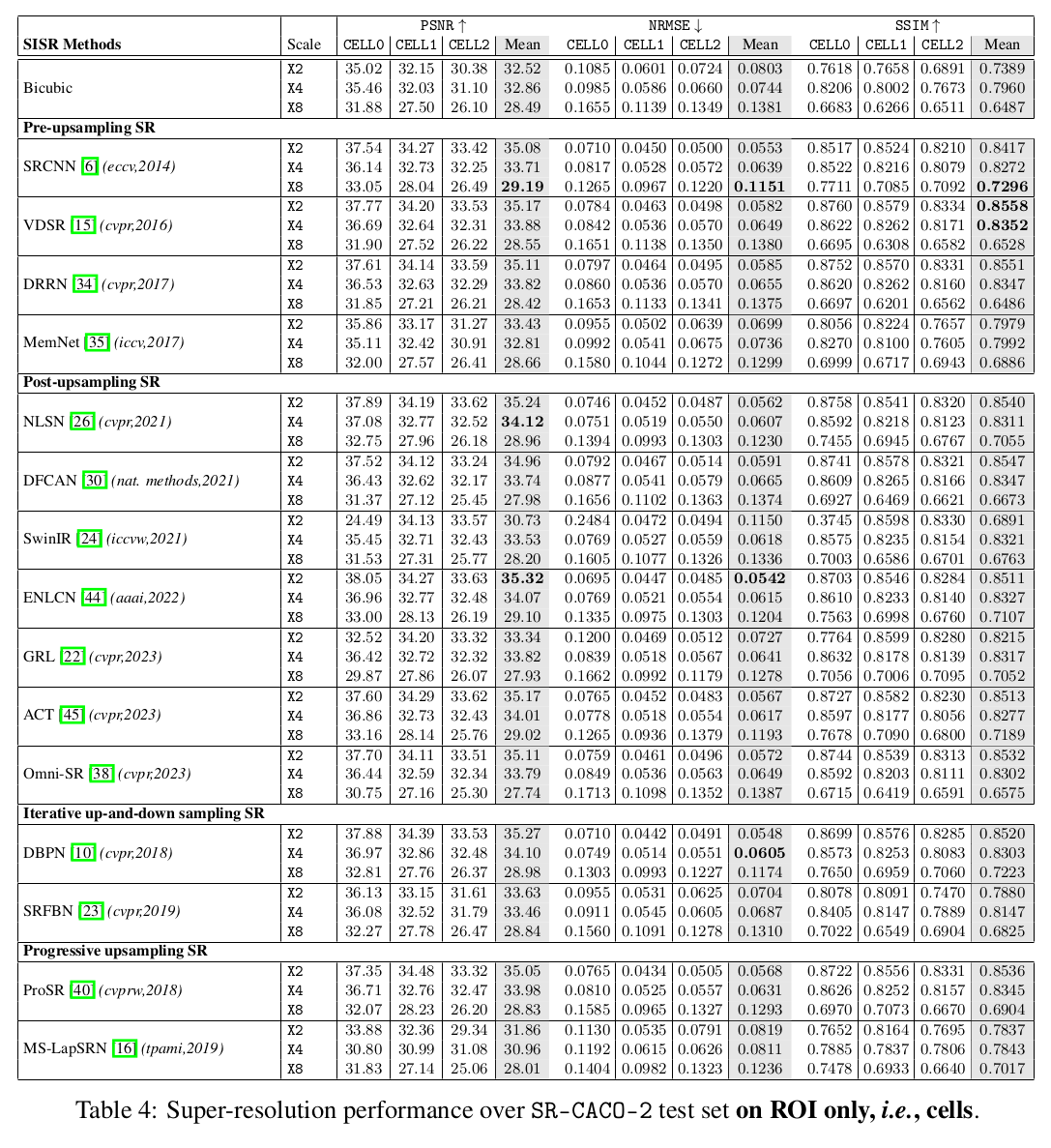
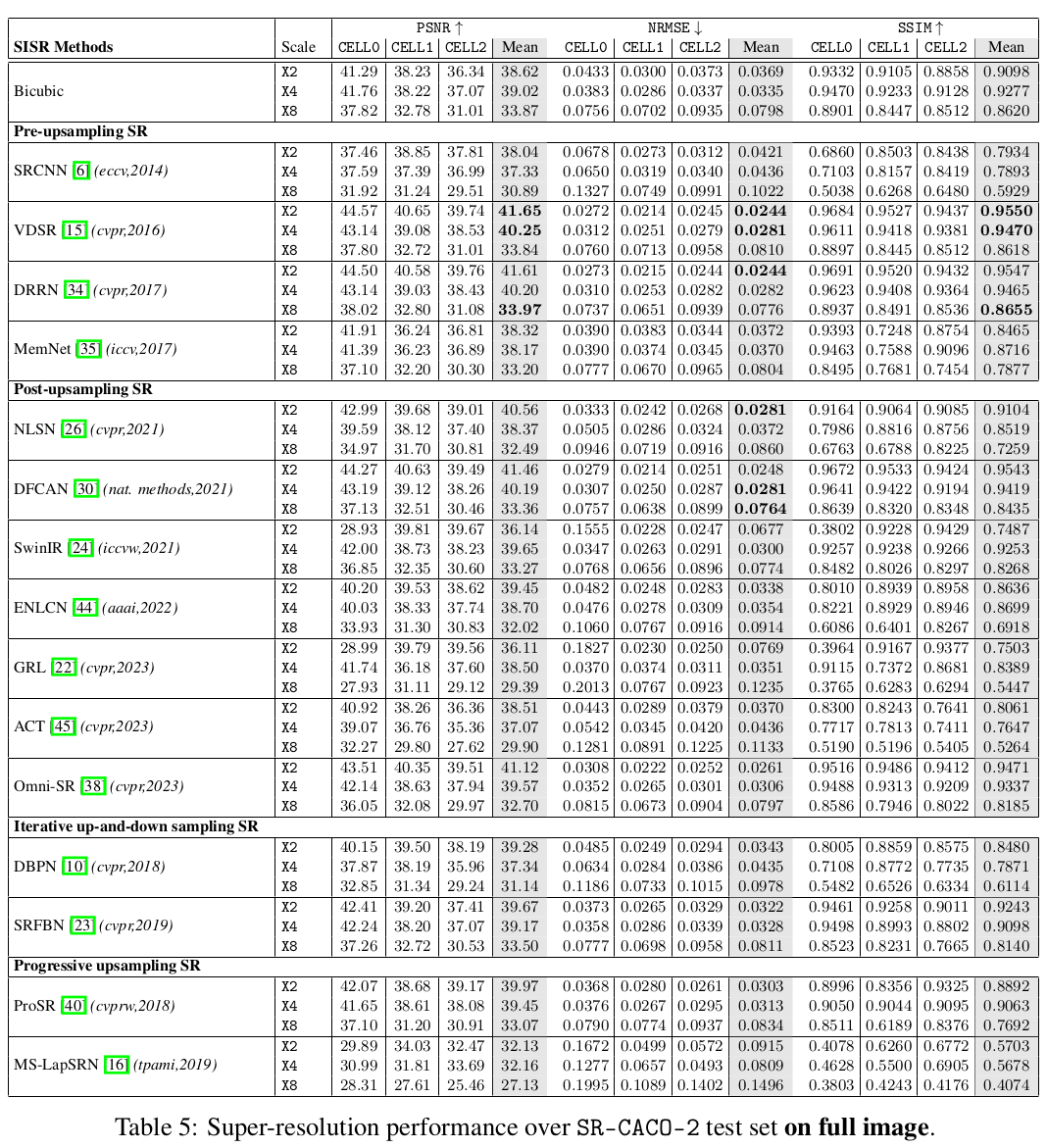
In this paper, we introduce a large scanning confocal microscopy dataset named
SR-CACO-2 that is comprised of low- and high-resolution image pairs marked for
three different fluorescent markers. It allows the evaluation of performance of
SISR methods on three different upscaling levels (X2, X4, X8). SR-CACO-2
contains the human epithelial cell line Caco-2 (ATCC HTB-37), and it is
composed of 22 tiles that have been translated in the form of 9,937 image
patches for experiments with SISR methods. Given the new SR-CACO-2 dataset,
we also provide benchmarking results for 15 state-of-the-art methods that are
representative of the main SISR families. Results show that these methods have
limited success in producing high-resolution textures, indicating that SR-CACO-2
represents a challenging problem. Our dataset, code and pretrained weights are
available: https://github.com/sbelharbi/sr-caco-2.
**Code: Pytorch 2.0.0**
## Citation:
```
@inproceedings{belharbi24-sr-caco-2,
title={SR-CACO-2: A Dataset for Confocal Fluorescence Microscopy Image Super-Resolution},
author={Belharbi, S. and Whitford, M.K.M. and Hoang, P. and Murtaza, S. and McCaffrey, L. and Granger, E.},
booktitle={NeurIPS},
year={2024}
}
```

## Pretrained weights (evaluation) :
We provide the weights for all the models (135 models: 15 methods x 3 cells
x 3 scales). Weights can be found at [Hugging Face](https://huggingface.co/sbelharbi/sr-caco-2) in the file [shared-trained-models.tar.gz](https://huggingface.co/sbelharbi/sr-caco-2/resolve/main/shared-trained-models.tar.gz?download=true).
The file [share-visualization-30-samples-test.zip](https://huggingface.co/sbelharbi/sr-caco-2/resolve/main/share-visualization-30-samples-test.zip?download=true) contains visual predictions on the test set.
The provided weights can be used to reproduce the reported results in the
paper in the paper: