Spaces:
Runtime error
Runtime error
Commit
·
c238491
1
Parent(s):
59ed1a3
Model files
Browse files- Attention_Extracter.py +47 -0
- EdgeWeightPredictorModel.py +39 -0
- GATv2DecoderModel.py +38 -0
- GATv2EncoderModel.py +32 -0
- GraphAnalysis.py +170 -0
- MultiOmicsGraphAttentionAutoencoderModel.py +155 -0
- OmicsConfig.py +27 -0
- app.py +0 -5
- data/README.md +1 -0
- data/survival.hnsc_data.csv +524 -0
- lc_models/MultiOmicsAutoencoder/trained_autoencoder/config.json +22 -0
- lc_models/MultiOmicsAutoencoder/trained_autoencoder/pytorch_model.bin +3 -0
- lc_models/MultiOmicsAutoencoder/trained_decoder/config.json +22 -0
- lc_models/MultiOmicsAutoencoder/trained_decoder/pytorch_model.bin +3 -0
- lc_models/MultiOmicsAutoencoder/trained_edge_weight_predictor/config.json +22 -0
- lc_models/MultiOmicsAutoencoder/trained_edge_weight_predictor/pytorch_model.bin +3 -0
- lc_models/MultiOmicsAutoencoder/trained_encoder/config.json +22 -0
- lc_models/MultiOmicsAutoencoder/trained_encoder/pytorch_model.bin +3 -0
- results/temp_k3_club_plan_meir.jpeg +0 -0
- results/temp_k3_club_plan_meir.png +0 -0
- results/temp_k5_plan_meir.jpeg +0 -0
- results/temp_k5_plan_meir.png +0 -0
- results/temp_median_survival.jpeg +0 -0
- results/temp_median_survival.png +0 -0
- train.py +105 -0
Attention_Extracter.py
ADDED
|
@@ -0,0 +1,47 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
import pickle
|
| 3 |
+
import numpy as np
|
| 4 |
+
|
| 5 |
+
class Attention_Extracter:
|
| 6 |
+
def __init__(self, graph_data_dict_path, encoder_model, gpu=False):
|
| 7 |
+
self.torch_device = 'cuda' if gpu else 'cpu'
|
| 8 |
+
|
| 9 |
+
self.graph_data_dict = torch.load(graph_data_dict_path)
|
| 10 |
+
self.encoder_model = encoder_model
|
| 11 |
+
self.encoder_model.to(self.torch_device)
|
| 12 |
+
self.encoder_model.eval()
|
| 13 |
+
self.latent_feat_dict, self.attention_scores1 = self.extract_latent_attention_features()
|
| 14 |
+
|
| 15 |
+
def extract_latent_attention_features(self):
|
| 16 |
+
latent_features = {}
|
| 17 |
+
attention_scores1 = {}
|
| 18 |
+
|
| 19 |
+
with torch.no_grad():
|
| 20 |
+
for graph_id, data in self.graph_data_dict.items():
|
| 21 |
+
data = data.to(self.torch_device)
|
| 22 |
+
z, attention_weights = self.encoder_model(data.x, data.edge_index, data.edge_attr)
|
| 23 |
+
latent_features[graph_id] = z.cpu()
|
| 24 |
+
|
| 25 |
+
# Handling the case where attention_weights is a tuple or other data structure
|
| 26 |
+
if isinstance(attention_weights, (list, tuple)):
|
| 27 |
+
attention_scores1[graph_id] = [aw for aw in attention_weights]
|
| 28 |
+
else:
|
| 29 |
+
attention_scores1[graph_id] = attention_weights.cpu()
|
| 30 |
+
|
| 31 |
+
return latent_features, attention_scores1
|
| 32 |
+
|
| 33 |
+
def load_edge_indices(self, glist_path, edge_matrix_path):
|
| 34 |
+
with open(glist_path, 'rb') as f:
|
| 35 |
+
glist = pickle.load(f)
|
| 36 |
+
|
| 37 |
+
edge_matrix = np.load(edge_matrix_path)
|
| 38 |
+
edge_matrix = torch.tensor(edge_matrix, dtype=torch.float)
|
| 39 |
+
edge_index = torch.nonzero(edge_matrix, as_tuple=False).t().contiguous()
|
| 40 |
+
edge_indices_dict = {}
|
| 41 |
+
|
| 42 |
+
for i in range(edge_index.shape[1]):
|
| 43 |
+
index1, index2 = edge_index[0, i].item(), edge_index[1, i].item()
|
| 44 |
+
gene1, gene2 = glist[index1], glist[index2]
|
| 45 |
+
edge_indices_dict[(index1, index2)] = (gene1, gene2)
|
| 46 |
+
|
| 47 |
+
return edge_indices_dict
|
EdgeWeightPredictorModel.py
ADDED
|
@@ -0,0 +1,39 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from transformers import PreTrainedModel
|
| 2 |
+
from OmicsConfig import OmicsConfig
|
| 3 |
+
from transformers import PretrainedConfig, PreTrainedModel
|
| 4 |
+
import torch
|
| 5 |
+
import torch.nn as nn
|
| 6 |
+
import torch.nn.functional as F
|
| 7 |
+
from torch_geometric.nn import GATv2Conv
|
| 8 |
+
from torch_geometric.data import Batch
|
| 9 |
+
from torch.utils.data import DataLoader
|
| 10 |
+
from torch.optim import AdamW
|
| 11 |
+
from torch_geometric.utils import negative_sampling
|
| 12 |
+
from torch.nn.functional import cosine_similarity
|
| 13 |
+
from torch.optim.lr_scheduler import StepLR
|
| 14 |
+
|
| 15 |
+
|
| 16 |
+
class EdgeWeightPredictorModel(PreTrainedModel):
|
| 17 |
+
config_class = OmicsConfig
|
| 18 |
+
base_model_prefix = "edge_weight_predictor"
|
| 19 |
+
|
| 20 |
+
def __init__(self, config):
|
| 21 |
+
super().__init__(config)
|
| 22 |
+
layers = []
|
| 23 |
+
input_size = 2 * config.out_channels
|
| 24 |
+
for hidden_size, activation in zip(config.edge_decoder_hidden_sizes, config.edge_decoder_activations):
|
| 25 |
+
layers.append(nn.Linear(input_size, hidden_size))
|
| 26 |
+
if activation == 'ReLU':
|
| 27 |
+
layers.append(nn.ReLU())
|
| 28 |
+
elif activation == 'Sigmoid':
|
| 29 |
+
layers.append(nn.Sigmoid())
|
| 30 |
+
elif activation == 'Tanh':
|
| 31 |
+
layers.append(nn.Tanh())
|
| 32 |
+
# Add more activations if needed
|
| 33 |
+
input_size = hidden_size
|
| 34 |
+
layers.append(nn.Linear(input_size, 1))
|
| 35 |
+
self.predictor = nn.Sequential(*layers)
|
| 36 |
+
|
| 37 |
+
def forward(self, z, edge_index):
|
| 38 |
+
edge_embeddings = torch.cat([z[edge_index[0]], z[edge_index[1]]], dim=-1)
|
| 39 |
+
return self.predictor(edge_embeddings)
|
GATv2DecoderModel.py
ADDED
|
@@ -0,0 +1,38 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from transformers import PreTrainedModel
|
| 2 |
+
from OmicsConfig import OmicsConfig
|
| 3 |
+
from transformers import PretrainedConfig, PreTrainedModel
|
| 4 |
+
import torch
|
| 5 |
+
import torch.nn as nn
|
| 6 |
+
import torch.nn.functional as F
|
| 7 |
+
from torch_geometric.nn import GATv2Conv
|
| 8 |
+
from torch_geometric.data import Batch
|
| 9 |
+
from torch.utils.data import DataLoader
|
| 10 |
+
from torch.optim import AdamW
|
| 11 |
+
from torch_geometric.utils import negative_sampling
|
| 12 |
+
from torch.nn.functional import cosine_similarity
|
| 13 |
+
from torch.optim.lr_scheduler import StepLR
|
| 14 |
+
|
| 15 |
+
from EdgeWeightPredictorModel import EdgeWeightPredictorModel
|
| 16 |
+
|
| 17 |
+
class GATv2DecoderModel(PreTrainedModel):
|
| 18 |
+
config_class = OmicsConfig
|
| 19 |
+
base_model_prefix = "gatv2_decoder"
|
| 20 |
+
|
| 21 |
+
def __init__(self, config):
|
| 22 |
+
super().__init__(config)
|
| 23 |
+
self.layers = nn.ModuleList([
|
| 24 |
+
nn.Linear(config.out_channels if i == 0 else config.out_channels, config.out_channels)
|
| 25 |
+
for i in range(config.num_layers)
|
| 26 |
+
])
|
| 27 |
+
self.fc = nn.Linear(config.out_channels, config.original_feature_size)
|
| 28 |
+
self.edge_weight_predictor = EdgeWeightPredictorModel(config)
|
| 29 |
+
|
| 30 |
+
def forward(self, z):
|
| 31 |
+
for layer in self.layers:
|
| 32 |
+
z = layer(z)
|
| 33 |
+
z = F.relu(z)
|
| 34 |
+
x_reconstructed = self.fc(z)
|
| 35 |
+
return x_reconstructed
|
| 36 |
+
|
| 37 |
+
def predict_edge_weights(self, z, edge_index):
|
| 38 |
+
return self.edge_weight_predictor(z, edge_index)
|
GATv2EncoderModel.py
ADDED
|
@@ -0,0 +1,32 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from transformers import PreTrainedModel
|
| 2 |
+
from OmicsConfig import OmicsConfig
|
| 3 |
+
from transformers import PretrainedConfig, PreTrainedModel
|
| 4 |
+
import torch
|
| 5 |
+
import torch.nn as nn
|
| 6 |
+
import torch.nn.functional as F
|
| 7 |
+
from torch_geometric.nn import GATv2Conv
|
| 8 |
+
from torch_geometric.data import Batch
|
| 9 |
+
from torch.utils.data import DataLoader
|
| 10 |
+
from torch.optim import AdamW
|
| 11 |
+
from torch_geometric.utils import negative_sampling
|
| 12 |
+
from torch.nn.functional import cosine_similarity
|
| 13 |
+
from torch.optim.lr_scheduler import StepLR
|
| 14 |
+
|
| 15 |
+
|
| 16 |
+
class GATv2EncoderModel(PreTrainedModel):
|
| 17 |
+
config_class = OmicsConfig
|
| 18 |
+
base_model_prefix = "gatv2_encoder"
|
| 19 |
+
|
| 20 |
+
def __init__(self, config):
|
| 21 |
+
super().__init__(config)
|
| 22 |
+
self.layers = nn.ModuleList([
|
| 23 |
+
GATv2Conv(config.in_channels if i == 0 else config.out_channels, config.out_channels, heads=1, concat=True, edge_dim=config.edge_attr_channels, add_self_loops=False)
|
| 24 |
+
for i in range(config.num_layers)
|
| 25 |
+
])
|
| 26 |
+
|
| 27 |
+
def forward(self, x, edge_index, edge_attr):
|
| 28 |
+
attention_weights = []
|
| 29 |
+
for layer in self.layers:
|
| 30 |
+
x, attn_weights = layer(x, edge_index, edge_attr, return_attention_weights=True)
|
| 31 |
+
attention_weights.append(attn_weights)
|
| 32 |
+
return x, attention_weights
|
GraphAnalysis.py
ADDED
|
@@ -0,0 +1,170 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import numpy as np
|
| 2 |
+
from sklearn.cluster import KMeans
|
| 3 |
+
from sklearn.decomposition import PCA
|
| 4 |
+
from sklearn.manifold import TSNE
|
| 5 |
+
from lifelines.statistics import logrank_test
|
| 6 |
+
from itertools import combinations
|
| 7 |
+
import matplotlib.pyplot as plt
|
| 8 |
+
from yellowbrick.cluster import KElbowVisualizer
|
| 9 |
+
import pandas as pd
|
| 10 |
+
import seaborn as sns
|
| 11 |
+
from lifelines import KaplanMeierFitter
|
| 12 |
+
import matplotlib.cm as cm
|
| 13 |
+
import itertools
|
| 14 |
+
import torch
|
| 15 |
+
|
| 16 |
+
class GraphAnalysis:
|
| 17 |
+
def __init__(self, EXTRACTER):
|
| 18 |
+
self.extracter = EXTRACTER
|
| 19 |
+
self.process()
|
| 20 |
+
|
| 21 |
+
def process(self):
|
| 22 |
+
latent_features_list = list(self.extracter.latent_feat_dict.values())
|
| 23 |
+
patient_list = list(self.extracter.latent_feat_dict.keys())
|
| 24 |
+
latentF = torch.stack(latent_features_list, dim=0)
|
| 25 |
+
self.latentF = np.squeeze(latentF.numpy())
|
| 26 |
+
self.pIDs = patient_list
|
| 27 |
+
self.df = pd.DataFrame(columns=['PC1','PC2','tX','tY','groups'], index=self.pIDs)
|
| 28 |
+
self.clnc_df = pd.read_csv('./data/survival.hnsc_data.csv').set_index('PatientID')
|
| 29 |
+
self.df = self.df.join(self.clnc_df)
|
| 30 |
+
|
| 31 |
+
def pca_tsne(self):
|
| 32 |
+
pca = PCA(n_components=2)
|
| 33 |
+
X_pca = pca.fit_transform(self.latentF)
|
| 34 |
+
self.df['PC1'] = X_pca[:,0]
|
| 35 |
+
self.df['PC2'] = X_pca[:,1]
|
| 36 |
+
tsne = TSNE(n_components=2)
|
| 37 |
+
X_tsne = tsne.fit_transform(self.latentF)
|
| 38 |
+
self.df['tX'] = X_tsne[:,0]
|
| 39 |
+
self.df['tY'] = X_tsne[:,1]
|
| 40 |
+
|
| 41 |
+
def find_optimal_clusters(self, min_clusters=2, max_clusters=11, save_path='./results/kelbow'):
|
| 42 |
+
model = KMeans(random_state=42)
|
| 43 |
+
visualizer = KElbowVisualizer(model, k=(min_clusters, max_clusters))
|
| 44 |
+
visualizer.fit(self.latentF)
|
| 45 |
+
visualizer.show()
|
| 46 |
+
fig = visualizer.ax.get_figure()
|
| 47 |
+
fig.savefig(save_path + ".png", dpi=150)
|
| 48 |
+
fig.savefig(save_path + ".jpeg", format="jpeg", dpi=150)
|
| 49 |
+
self.optimal_clusters = visualizer.elbow_value_
|
| 50 |
+
|
| 51 |
+
def cluster_data(self):
|
| 52 |
+
if self.optimal_clusters is None:
|
| 53 |
+
raise ValueError("Please run 'find_optimal_clusters' method before clustering the data.")
|
| 54 |
+
kmeans = KMeans(n_clusters=self.optimal_clusters, random_state=0).fit(self.latentF)
|
| 55 |
+
self.labels = kmeans.labels_
|
| 56 |
+
self.df['groups'] = self.labels
|
| 57 |
+
self.generate_color_list_based_on_median_survival()
|
| 58 |
+
|
| 59 |
+
def cluster_data2(self, kclust):
|
| 60 |
+
kmeans = KMeans(n_clusters=kclust, random_state=0).fit(self.latentF)
|
| 61 |
+
self.labels = kmeans.labels_
|
| 62 |
+
self.df['groups'] = self.labels
|
| 63 |
+
self.generate_color_list_based_on_median_survival()
|
| 64 |
+
|
| 65 |
+
def visualize_clusters(self):
|
| 66 |
+
plt.figure(figsize=(20,8))
|
| 67 |
+
plt.subplot(1,2,1)
|
| 68 |
+
sns.scatterplot(data=self.df, x='PC1', y='PC2', hue='groups', palette=self.color_list)
|
| 69 |
+
plt.subplot(1,2,2)
|
| 70 |
+
sns.scatterplot(data=self.df, x='tX', y='tY', hue='groups', palette=self.color_list)
|
| 71 |
+
|
| 72 |
+
def save_visualize_clusters(self):
|
| 73 |
+
plt.figure(figsize=(10,8))
|
| 74 |
+
sns.scatterplot(data=self.df, x='PC1', y='PC2', hue='groups', palette=self.color_list)
|
| 75 |
+
plt.savefig('./results/temp_pca.jpeg', dpi=300)
|
| 76 |
+
plt.savefig('./results/temp_pca.png', dpi=300)
|
| 77 |
+
plt.close()
|
| 78 |
+
plt.figure(figsize=(10,8))
|
| 79 |
+
sns.scatterplot(data=self.df, x='tX', y='tY', hue='groups', palette=self.color_list)
|
| 80 |
+
plt.savefig('./results/temp_tsne.jpeg', dpi=300)
|
| 81 |
+
plt.savefig('./results/temp_tsne.png', dpi=300)
|
| 82 |
+
|
| 83 |
+
def map_group_to_color(group):
|
| 84 |
+
return self.color_list[group]
|
| 85 |
+
|
| 86 |
+
def generate_color_list_based_on_median_survival(self):
|
| 87 |
+
groups = self.df['groups'].unique()
|
| 88 |
+
median_survival_times = {group: self.df[self.df['groups'] == group]['Overall Survival (Months)'].median() for group in groups}
|
| 89 |
+
sorted_groups = sorted(groups, key=median_survival_times.get, reverse=True)
|
| 90 |
+
vibgyor_colors = cm.rainbow(np.linspace(0, 1, len(groups)))
|
| 91 |
+
self.color_list = {group: color for group, color in zip(sorted_groups, vibgyor_colors)}
|
| 92 |
+
|
| 93 |
+
def perform_log_rank_test(self, alpha=0.05):
|
| 94 |
+
if self.df is None:
|
| 95 |
+
raise ValueError("Please run 'cluster_data' or 'cluster_data2' method before performing log rank test.")
|
| 96 |
+
groups = self.df['groups'].unique()
|
| 97 |
+
significant_pairs = []
|
| 98 |
+
for pair in itertools.combinations(groups, 2):
|
| 99 |
+
group_a = self.df[self.df['groups'] == pair[0]]
|
| 100 |
+
group_b = self.df[self.df['groups'] == pair[1]]
|
| 101 |
+
results = logrank_test(group_a['Overall Survival (Months)'], group_b['Overall Survival (Months)'], group_a['Overall Survival Status'], group_b['Overall Survival Status'])
|
| 102 |
+
if results.p_value < alpha:
|
| 103 |
+
significant_pairs.append(pair)
|
| 104 |
+
self.significant_pairs = significant_pairs
|
| 105 |
+
return self.significant_pairs
|
| 106 |
+
|
| 107 |
+
def generate_summary_table(self):
|
| 108 |
+
groups = self.df['groups'].unique()
|
| 109 |
+
summary_table = pd.DataFrame(columns=['Total number of patients', 'Alive', 'Deceased', 'Median survival time'], index=groups)
|
| 110 |
+
for group in groups:
|
| 111 |
+
group_data = self.df[self.df['groups'] == group]
|
| 112 |
+
total_patients = len(group_data)
|
| 113 |
+
alive = len(group_data[group_data['Overall Survival Status'] == 0])
|
| 114 |
+
deceased = len(group_data[group_data['Overall Survival Status'] == 1])
|
| 115 |
+
kmf = KaplanMeierFitter()
|
| 116 |
+
kmf.fit(group_data['Overall Survival (Months)'], group_data['Overall Survival Status'])
|
| 117 |
+
median_survival_time = kmf.median_survival_time_
|
| 118 |
+
summary_table.loc[group] = [total_patients, alive, deceased, median_survival_time]
|
| 119 |
+
return summary_table
|
| 120 |
+
|
| 121 |
+
def plot_kaplan_meier(self, plot_for_groups=True, name='temp_k5'):
|
| 122 |
+
kmf = KaplanMeierFitter()
|
| 123 |
+
plt.figure(figsize=(8, 6))
|
| 124 |
+
plt.grid(False)
|
| 125 |
+
if plot_for_groups:
|
| 126 |
+
groups = sorted(self.df['groups'].unique())
|
| 127 |
+
for i, group in enumerate(groups):
|
| 128 |
+
group_data = self.df[self.df['groups'] == group]
|
| 129 |
+
kmf.fit(group_data['Overall Survival (Months)'], group_data['Overall Survival Status'], label=f'Group {group}')
|
| 130 |
+
kmf.plot(ci_show=False, linewidth=2, color=self.color_list[group])
|
| 131 |
+
plt.title("Kaplan-Meier Curves for Each Group")
|
| 132 |
+
else:
|
| 133 |
+
kmf.fit(self.df['Overall Survival (Months)'], self.df['Overall Survival Status'], label='All Data')
|
| 134 |
+
kmf.plot(ci_show=False, linewidth=2, color='black')
|
| 135 |
+
plt.title("Kaplan-Meier Curve for All Data")
|
| 136 |
+
plt.gca().set_facecolor('#f5f5f5')
|
| 137 |
+
plt.grid(color='lightgrey', linestyle='-', linewidth=0.5)
|
| 138 |
+
plt.xlabel("Overall Survival (Months)", fontweight='bold')
|
| 139 |
+
plt.ylabel("Survival Probability", fontweight='bold')
|
| 140 |
+
plt.legend()
|
| 141 |
+
plt.savefig('./results/{}_plan_meir.jpeg'.format(name), dpi=300)
|
| 142 |
+
plt.savefig('./results/{}_plan_meir.png'.format(name), dpi=300)
|
| 143 |
+
plt.show()
|
| 144 |
+
|
| 145 |
+
def club_two_groups(self, primary_group, secondary_group):
|
| 146 |
+
self.df.loc[self.df['groups'] == secondary_group, 'groups'] = primary_group
|
| 147 |
+
unique_groups = sorted(self.df['groups'].unique())
|
| 148 |
+
mapping = {old: new for new, old in enumerate(unique_groups)}
|
| 149 |
+
self.df['groups'] = self.df['groups'].map(mapping)
|
| 150 |
+
self.generate_color_list_based_on_median_survival()
|
| 151 |
+
self.summary_table = self.generate_summary_table()
|
| 152 |
+
|
| 153 |
+
def plot_median_survival_bar(self, name='temp_k5'):
|
| 154 |
+
summary_df = self.generate_summary_table()
|
| 155 |
+
summary_df['group'] = summary_df.index
|
| 156 |
+
max_val = summary_df["Median survival time"].replace(np.inf, np.nan).max()
|
| 157 |
+
summary_df["Display Median"] = summary_df["Median survival time"].replace(np.inf, max_val * 1.1)
|
| 158 |
+
summary_df = summary_df.sort_index()
|
| 159 |
+
colors = [self.color_list[group] for group in summary_df.index]
|
| 160 |
+
num_groups = len(summary_df)
|
| 161 |
+
plt.figure(figsize=(6, num_groups * 0.8))
|
| 162 |
+
plt.grid(False)
|
| 163 |
+
sns.barplot(data=summary_df, y='group', x="Display Median", palette=colors, orient="h", order=summary_df.index)
|
| 164 |
+
plt.xlabel("Median Survival Time (Months)")
|
| 165 |
+
plt.ylabel("Groups")
|
| 166 |
+
plt.title("Median Survival Time by Group")
|
| 167 |
+
plt.tight_layout()
|
| 168 |
+
plt.savefig('./results/{}_median_survival.jpeg'.format(name), dpi=300)
|
| 169 |
+
plt.savefig('./results/{}_median_survival.png'.format(name), dpi=300)
|
| 170 |
+
plt.show()
|
MultiOmicsGraphAttentionAutoencoderModel.py
ADDED
|
@@ -0,0 +1,155 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from transformers import PreTrainedModel
|
| 2 |
+
from OmicsConfig import OmicsConfig
|
| 3 |
+
from transformers import PretrainedConfig, PreTrainedModel
|
| 4 |
+
import torch
|
| 5 |
+
import torch.nn as nn
|
| 6 |
+
import torch.nn.functional as F
|
| 7 |
+
from torch_geometric.nn import GATv2Conv
|
| 8 |
+
from torch_geometric.data import Batch
|
| 9 |
+
from torch.utils.data import DataLoader
|
| 10 |
+
from torch.optim import AdamW
|
| 11 |
+
from torch_geometric.utils import negative_sampling
|
| 12 |
+
from torch.nn.functional import cosine_similarity
|
| 13 |
+
from torch.optim.lr_scheduler import StepLR
|
| 14 |
+
|
| 15 |
+
from GATv2EncoderModel import GATv2EncoderModel
|
| 16 |
+
from GATv2DecoderModel import GATv2DecoderModel
|
| 17 |
+
from EdgeWeightPredictorModel import EdgeWeightPredictorModel
|
| 18 |
+
|
| 19 |
+
|
| 20 |
+
class MultiOmicsGraphAttentionAutoencoderModel(PreTrainedModel):
|
| 21 |
+
config_class = OmicsConfig
|
| 22 |
+
base_model_prefix = "graph-attention-autoencoder"
|
| 23 |
+
|
| 24 |
+
def __init__(self, config):
|
| 25 |
+
super().__init__(config)
|
| 26 |
+
self.encoder = GATv2EncoderModel(config)
|
| 27 |
+
self.decoder = GATv2DecoderModel(config)
|
| 28 |
+
self.optimizer = AdamW(list(self.encoder.parameters()) + list(self.decoder.parameters()), lr=config.learning_rate)
|
| 29 |
+
self.scheduler = StepLR(self.optimizer, step_size=30, gamma=0.7)
|
| 30 |
+
|
| 31 |
+
def forward(self, x, edge_index, edge_attr):
|
| 32 |
+
z, attention_weights = self.encoder(x, edge_index, edge_attr)
|
| 33 |
+
x_reconstructed = self.decoder(z)
|
| 34 |
+
return x_reconstructed, attention_weights
|
| 35 |
+
|
| 36 |
+
def predict_edge_weights(self, z, edge_index):
|
| 37 |
+
return self.decoder.predict_edge_weights(z, edge_index)
|
| 38 |
+
|
| 39 |
+
def train_model(self, data_loader, device):
|
| 40 |
+
self.encoder.to(device)
|
| 41 |
+
self.decoder.to(device)
|
| 42 |
+
self.encoder.train()
|
| 43 |
+
self.decoder.train()
|
| 44 |
+
total_loss = 0
|
| 45 |
+
total_cosine_similarity = 0
|
| 46 |
+
loss_weight_node = 1.0
|
| 47 |
+
loss_weight_edge = 1.0
|
| 48 |
+
loss_weight_edge_attr = 1.0
|
| 49 |
+
|
| 50 |
+
for data in data_loader:
|
| 51 |
+
data = data.to(device)
|
| 52 |
+
self.optimizer.zero_grad()
|
| 53 |
+
z, attention_weights = self.encoder(data.x, data.edge_index, data.edge_attr)
|
| 54 |
+
x_reconstructed = self.decoder(z)
|
| 55 |
+
node_loss = graph_reconstruction_loss(x_reconstructed, data.x)
|
| 56 |
+
edge_loss = edge_reconstruction_loss(z, data.edge_index)
|
| 57 |
+
cos_sim = cosine_similarity(x_reconstructed, data.x, dim=-1).mean()
|
| 58 |
+
total_cosine_similarity += cos_sim.item()
|
| 59 |
+
pred_edge_weights = self.decoder.predict_edge_weights(z, data.edge_index)
|
| 60 |
+
edge_weight_loss = edge_weight_reconstruction_loss(pred_edge_weights, data.edge_attr)
|
| 61 |
+
loss = (loss_weight_node * node_loss) + (loss_weight_edge * edge_loss) + (loss_weight_edge_attr * edge_weight_loss)
|
| 62 |
+
print(f"node_loss: {node_loss}, edge_loss: {edge_loss:.4f}, edge_weight_loss: {edge_weight_loss:.4f}, cosine_similarity: {cos_sim:.4f}")
|
| 63 |
+
loss.backward()
|
| 64 |
+
self.optimizer.step()
|
| 65 |
+
total_loss += loss.item()
|
| 66 |
+
|
| 67 |
+
avg_loss, avg_cosine_similarity = total_loss / len(data_loader), total_cosine_similarity / len(data_loader)
|
| 68 |
+
return avg_loss, avg_cosine_similarity
|
| 69 |
+
|
| 70 |
+
def fit(self, train_loader, validation_loader, epochs, device):
|
| 71 |
+
train_losses = []
|
| 72 |
+
val_losses = []
|
| 73 |
+
|
| 74 |
+
for epoch in range(1, epochs + 1):
|
| 75 |
+
train_loss, train_cosine_similarity = self.train_model(train_loader, device)
|
| 76 |
+
torch.cuda.empty_cache()
|
| 77 |
+
val_loss, val_cosine_similarity = self.validate(validation_loader, device)
|
| 78 |
+
print(f"Epoch: {epoch}, Train Loss: {train_loss:.4f}, Train Cosine Similarity: {train_cosine_similarity:.4f}, Validation Loss: {val_loss:.4f}, Validation Cosine Similarity: {val_cosine_similarity:.4f}")
|
| 79 |
+
self.scheduler.step()
|
| 80 |
+
|
| 81 |
+
return train_losses, val_losses
|
| 82 |
+
|
| 83 |
+
def validate(self, validation_loader, device):
|
| 84 |
+
self.encoder.to(device)
|
| 85 |
+
self.decoder.to(device)
|
| 86 |
+
self.encoder.eval()
|
| 87 |
+
self.decoder.eval()
|
| 88 |
+
total_loss = 0
|
| 89 |
+
total_cosine_similarity = 0
|
| 90 |
+
|
| 91 |
+
with torch.no_grad():
|
| 92 |
+
for data in validation_loader:
|
| 93 |
+
data = data.to(device)
|
| 94 |
+
z, attention_weights = self.encoder(data.x, data.edge_index, data.edge_attr)
|
| 95 |
+
x_reconstructed = self.decoder(z)
|
| 96 |
+
node_loss = graph_reconstruction_loss(x_reconstructed, data.x)
|
| 97 |
+
edge_loss = edge_reconstruction_loss(z, data.edge_index)
|
| 98 |
+
cos_sim = cosine_similarity(x_reconstructed, data.x, dim=-1).mean()
|
| 99 |
+
total_cosine_similarity += cos_sim.item()
|
| 100 |
+
loss = node_loss + edge_loss
|
| 101 |
+
total_loss += loss.item()
|
| 102 |
+
|
| 103 |
+
avg_loss = total_loss / len(validation_loader)
|
| 104 |
+
avg_cosine_similarity = total_cosine_similarity / len(validation_loader)
|
| 105 |
+
return avg_loss, avg_cosine_similarity
|
| 106 |
+
|
| 107 |
+
def evaluate(self, test_loader, device):
|
| 108 |
+
self.encoder.to(device)
|
| 109 |
+
self.decoder.to(device)
|
| 110 |
+
self.encoder.eval()
|
| 111 |
+
self.decoder.eval()
|
| 112 |
+
total_loss = 0
|
| 113 |
+
total_accuracy = 0
|
| 114 |
+
|
| 115 |
+
with torch.no_grad():
|
| 116 |
+
for data in test_loader:
|
| 117 |
+
data = data.to(device)
|
| 118 |
+
z, attention_weights = self.encoder(data.x, data.edge_index, data.edge_attr)
|
| 119 |
+
x_reconstructed = self.decoder(z)
|
| 120 |
+
node_loss = graph_reconstruction_loss(x_reconstructed, data.x)
|
| 121 |
+
edge_loss = edge_reconstruction_loss(z, data.edge_index)
|
| 122 |
+
cos_sim = cosine_similarity(x_reconstructed, data.x, dim=-1).mean()
|
| 123 |
+
total_cosine_similarity += cos_sim.item()
|
| 124 |
+
loss = node_loss + edge_loss
|
| 125 |
+
total_loss += loss.item()
|
| 126 |
+
|
| 127 |
+
avg_loss = total_loss / len(validation_loader)
|
| 128 |
+
avg_cosine_similarity = total_cosine_similarity / len(validation_loader)
|
| 129 |
+
return avg_loss, avg_cosine_similarity
|
| 130 |
+
|
| 131 |
+
# Define a collate function for the DataLoader
|
| 132 |
+
def collate_graph_data(batch):
|
| 133 |
+
return Batch.from_data_list(batch)
|
| 134 |
+
|
| 135 |
+
# Define a function to create a DataLoader
|
| 136 |
+
def create_data_loader(train_data, batch_size=1, shuffle=True):
|
| 137 |
+
graph_data = list(train_data.values())
|
| 138 |
+
return DataLoader(graph_data, batch_size=batch_size, shuffle=shuffle, collate_fn=collate_graph_data)
|
| 139 |
+
|
| 140 |
+
# Define functions for the losses
|
| 141 |
+
def graph_reconstruction_loss(pred_features, true_features):
|
| 142 |
+
return F.mse_loss(pred_features, true_features)
|
| 143 |
+
|
| 144 |
+
def edge_reconstruction_loss(z, pos_edge_index, neg_edge_index=None):
|
| 145 |
+
pos_logits = (z[pos_edge_index[0]] * z[pos_edge_index[1]]).sum(dim=-1)
|
| 146 |
+
pos_loss = F.binary_cross_entropy_with_logits(pos_logits, torch.ones_like(pos_logits))
|
| 147 |
+
if neg_edge_index is None:
|
| 148 |
+
neg_edge_index = negative_sampling(pos_edge_index, z.size(0))
|
| 149 |
+
neg_logits = (z[neg_edge_index[0]] * z[neg_edge_index[1]]).sum(dim=-1)
|
| 150 |
+
neg_loss = F.binary_cross_entropy_with_logits(neg_logits, torch.zeros_like(neg_logits))
|
| 151 |
+
return pos_loss + neg_loss
|
| 152 |
+
|
| 153 |
+
def edge_weight_reconstruction_loss(pred_weights, true_weights):
|
| 154 |
+
pred_weights = pred_weights.squeeze(-1)
|
| 155 |
+
return F.mse_loss(pred_weights, true_weights)
|
OmicsConfig.py
ADDED
|
@@ -0,0 +1,27 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from transformers import PretrainedConfig
|
| 2 |
+
from transformers import PretrainedConfig, PreTrainedModel
|
| 3 |
+
import torch
|
| 4 |
+
import torch.nn as nn
|
| 5 |
+
import torch.nn.functional as F
|
| 6 |
+
from torch_geometric.nn import GATv2Conv
|
| 7 |
+
from torch_geometric.data import Batch
|
| 8 |
+
from torch.utils.data import DataLoader
|
| 9 |
+
from torch.optim import AdamW
|
| 10 |
+
from torch_geometric.utils import negative_sampling
|
| 11 |
+
from torch.nn.functional import cosine_similarity
|
| 12 |
+
from torch.optim.lr_scheduler import StepLR
|
| 13 |
+
|
| 14 |
+
|
| 15 |
+
class OmicsConfig(PretrainedConfig):
|
| 16 |
+
model_type = "omics-graph-network"
|
| 17 |
+
|
| 18 |
+
def __init__(self, in_channels=768, edge_attr_channels=128, out_channels=128, original_feature_size=768, learning_rate=0.01, num_layers=1, edge_decoder_hidden_sizes=[128], edge_decoder_activations=['ReLU'], **kwargs):
|
| 19 |
+
super().__init__(**kwargs)
|
| 20 |
+
self.in_channels = in_channels
|
| 21 |
+
self.edge_attr_channels = edge_attr_channels
|
| 22 |
+
self.out_channels = out_channels
|
| 23 |
+
self.original_feature_size = original_feature_size
|
| 24 |
+
self.learning_rate = learning_rate
|
| 25 |
+
self.num_layers = num_layers
|
| 26 |
+
self.edge_decoder_hidden_sizes = edge_decoder_hidden_sizes
|
| 27 |
+
self.edge_decoder_activations = edge_decoder_activations
|
app.py
CHANGED
|
@@ -14,11 +14,6 @@ from lifelines.statistics import logrank_test
|
|
| 14 |
import os
|
| 15 |
import subprocess
|
| 16 |
|
| 17 |
-
# Clone the GitHub repository
|
| 18 |
-
if not os.path.exists('/workspace/MultiOmics-Graph-Attention-Autoencoder'):
|
| 19 |
-
subprocess.run(['git', 'clone', 'https://github.com/VatsalPatel18/MultiOmics-Graph-Attention-Autoencoder.git', '/workspace/MultiOmics-Graph-Attention-Autoencoder'])
|
| 20 |
-
subprocess.run(['git', 'clone', 'https://huggingface.co/VatsalPatel18/HNSCC-MultiOmics-Graph-Attention-Autoencoder', '/workspace/HNSCC-MultiOmics-Graph-Attention-Autoencoder'])
|
| 21 |
-
|
| 22 |
from MultiOmicsGraphAttentionAutoencoderModel import MultiOmicsGraphAttentionAutoencoderModel
|
| 23 |
from OmicsConfig import OmicsConfig
|
| 24 |
from Attention_Extracter import Attention_Extracter
|
|
|
|
| 14 |
import os
|
| 15 |
import subprocess
|
| 16 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 17 |
from MultiOmicsGraphAttentionAutoencoderModel import MultiOmicsGraphAttentionAutoencoderModel
|
| 18 |
from OmicsConfig import OmicsConfig
|
| 19 |
from Attention_Extracter import Attention_Extracter
|
data/README.md
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
Data Here
|
data/survival.hnsc_data.csv
ADDED
|
@@ -0,0 +1,524 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
PatientID,Overall Survival Status,Overall Survival (Months)
|
| 2 |
+
TCGA-4P-AA8J-01,0,3.353387908
|
| 3 |
+
TCGA-BA-4074-01,1,15.18887464
|
| 4 |
+
TCGA-BA-4076-01,1,13.6436861
|
| 5 |
+
TCGA-BA-4078-01,1,9.073873163
|
| 6 |
+
TCGA-BA-5149-01,1,26.49833974
|
| 7 |
+
TCGA-BA-5151-01,0,23.73672617
|
| 8 |
+
TCGA-BA-5152-01,0,42.34474143
|
| 9 |
+
TCGA-BA-5153-01,1,57.92813229
|
| 10 |
+
TCGA-BA-5555-01,0,17.09570306
|
| 11 |
+
TCGA-BA-5556-01,0,23.83535523
|
| 12 |
+
TCGA-BA-5557-01,0,20.48196732
|
| 13 |
+
TCGA-BA-5558-01,0,65.58832232
|
| 14 |
+
TCGA-BA-5559-01,1,68.4814413
|
| 15 |
+
TCGA-BA-6868-01,1,15.51763816
|
| 16 |
+
TCGA-BA-6869-01,0,21.17237071
|
| 17 |
+
TCGA-BA-6870-01,1,14.82723477
|
| 18 |
+
TCGA-BA-6871-01,1,3.55064602
|
| 19 |
+
TCGA-BA-6872-01,1,12.62451918
|
| 20 |
+
TCGA-BA-6873-01,0,4.010914949
|
| 21 |
+
TCGA-BA-7269-01,0,41.85159615
|
| 22 |
+
TCGA-BA-A4IF-01,0,29.42433508
|
| 23 |
+
TCGA-BA-A4IG-01,0,28.10928099
|
| 24 |
+
TCGA-BA-A4IH-01,0,20.44909097
|
| 25 |
+
TCGA-BA-A4II-01,0,30.18049117
|
| 26 |
+
TCGA-BA-A6D8-01,0,27.94489923
|
| 27 |
+
TCGA-BA-A6DA-01,0,11.53959957
|
| 28 |
+
TCGA-BA-A6DB-01,0,7.101292041
|
| 29 |
+
TCGA-BA-A6DD-01,1,5.687608903
|
| 30 |
+
TCGA-BA-A6DE-01,0,14.4655949
|
| 31 |
+
TCGA-BA-A6DF-01,1,7.824571786
|
| 32 |
+
TCGA-BA-A6DG-01,1,2.268468291
|
| 33 |
+
TCGA-BA-A6DI-01,1,11.04645429
|
| 34 |
+
TCGA-BA-A6DJ-01,1,13.38067528
|
| 35 |
+
TCGA-BA-A6DL-01,0,20.48196732
|
| 36 |
+
TCGA-BA-A8YP-01,0,16.40529967
|
| 37 |
+
TCGA-BB-4217-01,0,6.147877832
|
| 38 |
+
TCGA-BB-4223-01,0,105.8947299
|
| 39 |
+
TCGA-BB-4224-01,0,9.139625867
|
| 40 |
+
TCGA-BB-4225-01,0,4.799947398
|
| 41 |
+
TCGA-BB-4227-01,0,4.405431173
|
| 42 |
+
TCGA-BB-4228-01,0,18.37788079
|
| 43 |
+
TCGA-BB-7861-01,0,22.42167209
|
| 44 |
+
TCGA-BB-7862-01,0,36.72288523
|
| 45 |
+
TCGA-BB-7863-01,0,33.69826084
|
| 46 |
+
TCGA-BB-7864-01,0,50.20218957
|
| 47 |
+
TCGA-BB-7866-01,0,44.97484959
|
| 48 |
+
TCGA-BB-7870-01,0,66.27872571
|
| 49 |
+
TCGA-BB-7871-01,0,24.65726403
|
| 50 |
+
TCGA-BB-7872-01,0,38.39957918
|
| 51 |
+
TCGA-BB-8596-01,0,71.04579676
|
| 52 |
+
TCGA-BB-8601-01,0,20.51484367
|
| 53 |
+
TCGA-BB-A5HU-01,0,25.7093073
|
| 54 |
+
TCGA-BB-A5HY-01,1,10.55330901
|
| 55 |
+
TCGA-BB-A5HZ-01,0,27.18874314
|
| 56 |
+
TCGA-BB-A6UM-01,0,12.92040635
|
| 57 |
+
TCGA-BB-A6UO-01,1,8.810862347
|
| 58 |
+
TCGA-C9-A47Z-01,1,6.27938324
|
| 59 |
+
TCGA-C9-A480-01,0,12.69027189
|
| 60 |
+
TCGA-CN-4722-01,0,48.75563008
|
| 61 |
+
TCGA-CN-4723-01,0,55.85692212
|
| 62 |
+
TCGA-CN-4725-01,0,38.03793931
|
| 63 |
+
TCGA-CN-4726-01,1,4.66844199
|
| 64 |
+
TCGA-CN-4727-01,0,51.28710918
|
| 65 |
+
TCGA-CN-4728-01,0,56.67883092
|
| 66 |
+
TCGA-CN-4729-01,0,12.88753
|
| 67 |
+
TCGA-CN-4730-01,0,26.85997962
|
| 68 |
+
TCGA-CN-4731-01,1,32.81059934
|
| 69 |
+
TCGA-CN-4733-01,0,52.14189434
|
| 70 |
+
TCGA-CN-4734-01,0,55.56103495
|
| 71 |
+
TCGA-CN-4735-01,0,57.10622349
|
| 72 |
+
TCGA-CN-4736-01,1,12.98615906
|
| 73 |
+
TCGA-CN-4737-01,0,20.54772003
|
| 74 |
+
TCGA-CN-4738-01,1,14.33408949
|
| 75 |
+
TCGA-CN-4739-01,1,45.82963474
|
| 76 |
+
TCGA-CN-4740-01,1,27.58325936
|
| 77 |
+
TCGA-CN-4741-01,0,73.61015222
|
| 78 |
+
TCGA-CN-4742-01,1,13.05191176
|
| 79 |
+
TCGA-CN-5355-01,0,42.01597791
|
| 80 |
+
TCGA-CN-5356-01,0,46.32278002
|
| 81 |
+
TCGA-CN-5358-01,1,8.580727882
|
| 82 |
+
TCGA-CN-5359-01,1,12.39438472
|
| 83 |
+
TCGA-CN-5360-01,0,71.30880758
|
| 84 |
+
TCGA-CN-5361-01,1,69.69786633
|
| 85 |
+
TCGA-CN-5363-01,1,8.317717066
|
| 86 |
+
TCGA-CN-5364-01,1,16.20804156
|
| 87 |
+
TCGA-CN-5365-01,1,11.53959957
|
| 88 |
+
TCGA-CN-5366-01,1,11.83548673
|
| 89 |
+
TCGA-CN-5367-01,1,11.57247592
|
| 90 |
+
TCGA-CN-5369-01,1,12.49301378
|
| 91 |
+
TCGA-CN-5370-01,1,8.514975178
|
| 92 |
+
TCGA-CN-5373-01,0,52.07614163
|
| 93 |
+
TCGA-CN-5374-01,1,56.94184173
|
| 94 |
+
TCGA-CN-6010-01,0,50.07068416
|
| 95 |
+
TCGA-CN-6011-01,0,30.67363645
|
| 96 |
+
TCGA-CN-6012-01,0,47.99947398
|
| 97 |
+
TCGA-CN-6013-01,1,23.90110793
|
| 98 |
+
TCGA-CN-6016-01,0,47.44057599
|
| 99 |
+
TCGA-CN-6017-01,1,28.04352829
|
| 100 |
+
TCGA-CN-6018-01,1,19.06828418
|
| 101 |
+
TCGA-CN-6019-01,0,34.12565342
|
| 102 |
+
TCGA-CN-6020-01,1,6.739652168
|
| 103 |
+
TCGA-CN-6021-01,1,9.073873163
|
| 104 |
+
TCGA-CN-6022-01,1,9.238254923
|
| 105 |
+
TCGA-CN-6023-01,0,52.07614163
|
| 106 |
+
TCGA-CN-6024-01,1,11.07933064
|
| 107 |
+
TCGA-CN-6988-01,0,10.45467995
|
| 108 |
+
TCGA-CN-6989-01,1,32.218825
|
| 109 |
+
TCGA-CN-6992-01,0,35.04619128
|
| 110 |
+
TCGA-CN-6994-01,0,38.89272446
|
| 111 |
+
TCGA-CN-6995-01,1,3.682151429
|
| 112 |
+
TCGA-CN-6996-01,1,17.42446658
|
| 113 |
+
TCGA-CN-6997-01,1,32.48183582
|
| 114 |
+
TCGA-CN-6998-01,1,11.73685768
|
| 115 |
+
TCGA-CN-A497-01,0,35.01331492
|
| 116 |
+
TCGA-CN-A498-01,1,25.41342013
|
| 117 |
+
TCGA-CN-A499-01,0,23.57234441
|
| 118 |
+
TCGA-CN-A49A-01,1,17.29296117
|
| 119 |
+
TCGA-CN-A49B-01,0,29.72022224
|
| 120 |
+
TCGA-CN-A49C-01,0,21.20524707
|
| 121 |
+
TCGA-CN-A63T-01,0,7.397179209
|
| 122 |
+
TCGA-CN-A63U-01,0,31.69280337
|
| 123 |
+
TCGA-CN-A63V-01,0,22.32304304
|
| 124 |
+
TCGA-CN-A63W-01,1,12.39438472
|
| 125 |
+
TCGA-CN-A63Y-01,0,16.56968143
|
| 126 |
+
TCGA-CN-A640-01,1,4.405431173
|
| 127 |
+
TCGA-CN-A641-01,0,12.0656212
|
| 128 |
+
TCGA-CN-A642-01,1,2.695860867
|
| 129 |
+
TCGA-CN-A6UY-01,0,23.44083901
|
| 130 |
+
TCGA-CN-A6V1-01,0,19.82444028
|
| 131 |
+
TCGA-CN-A6V3-01,0,24.39425321
|
| 132 |
+
TCGA-CN-A6V6-01,0,20.87648355
|
| 133 |
+
TCGA-CN-A6V7-01,0,19.52855311
|
| 134 |
+
TCGA-CQ-5323-01,0,48.19673209
|
| 135 |
+
TCGA-CQ-5324-01,0,52.3720288
|
| 136 |
+
TCGA-CQ-5325-01,1,21.50113423
|
| 137 |
+
TCGA-CQ-5326-01,1,2.925995332
|
| 138 |
+
TCGA-CQ-5327-01,0,54.57474439
|
| 139 |
+
TCGA-CQ-5329-01,0,70.45402242
|
| 140 |
+
TCGA-CQ-5330-01,0,62.36643982
|
| 141 |
+
TCGA-CQ-5331-01,0,45.9940165
|
| 142 |
+
TCGA-CQ-5332-01,1,10.4218036
|
| 143 |
+
TCGA-CQ-5333-01,1,11.21083605
|
| 144 |
+
TCGA-CQ-5334-01,1,4.241049413
|
| 145 |
+
TCGA-CQ-6218-01,0,41.19406911
|
| 146 |
+
TCGA-CQ-6219-01,1,15.74777263
|
| 147 |
+
TCGA-CQ-6220-01,1,32.38320676
|
| 148 |
+
TCGA-CQ-6222-01,0,66.27872571
|
| 149 |
+
TCGA-CQ-6223-01,0,46.94743071
|
| 150 |
+
TCGA-CQ-6224-01,0,56.58020186
|
| 151 |
+
TCGA-CQ-6225-01,1,13.24916987
|
| 152 |
+
TCGA-CQ-6227-01,1,4.241049413
|
| 153 |
+
TCGA-CQ-6228-01,1,14.99161653
|
| 154 |
+
TCGA-CQ-6229-01,0,38.76121906
|
| 155 |
+
TCGA-CQ-7063-01,0,70.1252589
|
| 156 |
+
TCGA-CQ-7064-01,0,64.86504258
|
| 157 |
+
TCGA-CQ-7065-01,0,53.52270112
|
| 158 |
+
TCGA-CQ-7067-01,0,16.73406319
|
| 159 |
+
TCGA-CQ-7068-01,0,43.03514482
|
| 160 |
+
TCGA-CQ-7069-01,0,41.8844725
|
| 161 |
+
TCGA-CQ-7071-01,0,43.10089752
|
| 162 |
+
TCGA-CQ-7072-01,0,77.55531446
|
| 163 |
+
TCGA-CQ-A4C6-01,0,44.48170431
|
| 164 |
+
TCGA-CQ-A4C9-01,0,23.24358089
|
| 165 |
+
TCGA-CQ-A4CA-01,0,
|
| 166 |
+
TCGA-CQ-A4CB-01,0,29.35858237
|
| 167 |
+
TCGA-CQ-A4CD-01,0,33.59963179
|
| 168 |
+
TCGA-CQ-A4CE-01,0,29.49008778
|
| 169 |
+
TCGA-CQ-A4CG-01,1,14.13683138
|
| 170 |
+
TCGA-CQ-A4CH-01,1,12.46013742
|
| 171 |
+
TCGA-CQ-A4CI-01,0,31.23253444
|
| 172 |
+
TCGA-CR-5243-01,0,84.22921393
|
| 173 |
+
TCGA-CR-5247-01,0,11.76973403
|
| 174 |
+
TCGA-CR-5248-01,0,54.67337344
|
| 175 |
+
TCGA-CR-5249-01,0,37.87355755
|
| 176 |
+
TCGA-CR-5250-01,0,26.26820528
|
| 177 |
+
TCGA-CR-6467-01,0,58.42127758
|
| 178 |
+
TCGA-CR-6470-01,0,50.00493145
|
| 179 |
+
TCGA-CR-6471-01,1,39.51737515
|
| 180 |
+
TCGA-CR-6472-01,0,34.52016964
|
| 181 |
+
TCGA-CR-6473-01,0,36.98589605
|
| 182 |
+
TCGA-CR-6474-01,1,18.54226255
|
| 183 |
+
TCGA-CR-6477-01,0,16.89844495
|
| 184 |
+
TCGA-CR-6478-01,1,6.016372423
|
| 185 |
+
TCGA-CR-6480-01,0,11.90123944
|
| 186 |
+
TCGA-CR-6481-01,0,10.22454548
|
| 187 |
+
TCGA-CR-6482-01,0,11.34234145
|
| 188 |
+
TCGA-CR-6484-01,0,11.63822862
|
| 189 |
+
TCGA-CR-6487-01,0,7.693066377
|
| 190 |
+
TCGA-CR-6488-01,0,12.46013742
|
| 191 |
+
TCGA-CR-6491-01,0,22.78331196
|
| 192 |
+
TCGA-CR-6492-01,0,15.74777263
|
| 193 |
+
TCGA-CR-6493-01,1,9.271131275
|
| 194 |
+
TCGA-CR-7364-01,0,47.17756518
|
| 195 |
+
TCGA-CR-7365-01,0,39.15573528
|
| 196 |
+
TCGA-CR-7367-01,0,47.34194694
|
| 197 |
+
TCGA-CR-7368-01,0,40.93105829
|
| 198 |
+
TCGA-CR-7369-01,1,35.83522372
|
| 199 |
+
TCGA-CR-7370-01,0,3.452016964
|
| 200 |
+
TCGA-CR-7371-01,1,3.090377092
|
| 201 |
+
TCGA-CR-7372-01,0,24.9531512
|
| 202 |
+
TCGA-CR-7373-01,0,29.22707696
|
| 203 |
+
TCGA-CR-7374-01,0,0.986290561
|
| 204 |
+
TCGA-CR-7376-01,0,31.95581418
|
| 205 |
+
TCGA-CR-7377-01,1,9.172502219
|
| 206 |
+
TCGA-CR-7379-01,0,34.05990071
|
| 207 |
+
TCGA-CR-7380-01,1,19.92306934
|
| 208 |
+
TCGA-CR-7382-01,0,26.16957622
|
| 209 |
+
TCGA-CR-7383-01,1,17.12857941
|
| 210 |
+
TCGA-CR-7385-01,0,32.77772298
|
| 211 |
+
TCGA-CR-7386-01,0,47.01318342
|
| 212 |
+
TCGA-CR-7388-01,1,27.05723773
|
| 213 |
+
TCGA-CR-7389-01,0,12.88753
|
| 214 |
+
TCGA-CR-7390-01,0,49.57753888
|
| 215 |
+
TCGA-CR-7391-01,0,30.01610941
|
| 216 |
+
TCGA-CR-7392-01,0,46.84880166
|
| 217 |
+
TCGA-CR-7393-01,0,32.64621758
|
| 218 |
+
TCGA-CR-7394-01,0,44.25156985
|
| 219 |
+
TCGA-CR-7395-01,0,30.5750074
|
| 220 |
+
TCGA-CR-7397-01,0,24.78876944
|
| 221 |
+
TCGA-CR-7398-01,0,5.128710918
|
| 222 |
+
TCGA-CR-7399-01,0,5.950619719
|
| 223 |
+
TCGA-CR-7401-01,0,35.40783115
|
| 224 |
+
TCGA-CR-7402-01,0,29.95035671
|
| 225 |
+
TCGA-CR-7404-01,0,48.3939902
|
| 226 |
+
TCGA-CV-5430-01,0,139.428609
|
| 227 |
+
TCGA-CV-5431-01,1,17.16145577
|
| 228 |
+
TCGA-CV-5432-01,0,129.2040635
|
| 229 |
+
TCGA-CV-5434-01,1,108.9522307
|
| 230 |
+
TCGA-CV-5435-01,1,76.24026038
|
| 231 |
+
TCGA-CV-5436-01,1,19.19978959
|
| 232 |
+
TCGA-CV-5439-01,1,17.95048821
|
| 233 |
+
TCGA-CV-5440-01,0,107.5056712
|
| 234 |
+
TCGA-CV-5441-01,0,94.88115199
|
| 235 |
+
TCGA-CV-5442-01,0,76.5032712
|
| 236 |
+
TCGA-CV-5443-01,0,91.52776408
|
| 237 |
+
TCGA-CV-5444-01,0,80.11966992
|
| 238 |
+
TCGA-CV-5966-01,1,17.91761186
|
| 239 |
+
TCGA-CV-5970-01,1,13.34779893
|
| 240 |
+
TCGA-CV-5971-01,0,23.04632278
|
| 241 |
+
TCGA-CV-5973-01,0,86.82644574
|
| 242 |
+
TCGA-CV-5976-01,0,48.59124832
|
| 243 |
+
TCGA-CV-5977-01,0,60.49248775
|
| 244 |
+
TCGA-CV-5978-01,1,7.068415689
|
| 245 |
+
TCGA-CV-5979-01,0,43.23240293
|
| 246 |
+
TCGA-CV-6003-01,0,54.73912615
|
| 247 |
+
TCGA-CV-6433-01,0,21.07374166
|
| 248 |
+
TCGA-CV-6436-01,0,62.43219252
|
| 249 |
+
TCGA-CV-6441-01,1,9.599894796
|
| 250 |
+
TCGA-CV-6933-01,1,90.11408094
|
| 251 |
+
TCGA-CV-6934-01,1,2.136962883
|
| 252 |
+
TCGA-CV-6935-01,1,9.698523852
|
| 253 |
+
TCGA-CV-6936-01,1,5.457474439
|
| 254 |
+
TCGA-CV-6937-01,1,20.51484367
|
| 255 |
+
TCGA-CV-6938-01,1,4.734194694
|
| 256 |
+
TCGA-CV-6939-01,1,21.89565046
|
| 257 |
+
TCGA-CV-6940-01,1,26.43258704
|
| 258 |
+
TCGA-CV-6941-01,1,11.2437124
|
| 259 |
+
TCGA-CV-6942-01,0,140.7765394
|
| 260 |
+
TCGA-CV-6943-01,1,19.79156393
|
| 261 |
+
TCGA-CV-6945-01,1,12.03274485
|
| 262 |
+
TCGA-CV-6948-01,1,42.37761778
|
| 263 |
+
TCGA-CV-6950-01,1,15.09024559
|
| 264 |
+
TCGA-CV-6951-01,1,30.08186212
|
| 265 |
+
TCGA-CV-6952-01,1,6.082125127
|
| 266 |
+
TCGA-CV-6953-01,1,53.9500937
|
| 267 |
+
TCGA-CV-6954-01,1,65.81845678
|
| 268 |
+
TCGA-CV-6955-01,1,10.98070158
|
| 269 |
+
TCGA-CV-6956-01,1,7.134168393
|
| 270 |
+
TCGA-CV-6959-01,1,8.416346122
|
| 271 |
+
TCGA-CV-6960-01,1,28.33941546
|
| 272 |
+
TCGA-CV-6961-01,1,2.498602755
|
| 273 |
+
TCGA-CV-6962-01,1,4.142420357
|
| 274 |
+
TCGA-CV-7089-01,1,64.83216622
|
| 275 |
+
TCGA-CV-7090-01,0,172.6666009
|
| 276 |
+
TCGA-CV-7091-01,0,111.1549463
|
| 277 |
+
TCGA-CV-7095-01,1,18.80527337
|
| 278 |
+
TCGA-CV-7097-01,1,12.65739554
|
| 279 |
+
TCGA-CV-7099-01,1,7.988953546
|
| 280 |
+
TCGA-CV-7100-01,1,9.008120459
|
| 281 |
+
TCGA-CV-7101-01,1,5.260216326
|
| 282 |
+
TCGA-CV-7102-01,1,1.841075714
|
| 283 |
+
TCGA-CV-7103-01,1,52.3062761
|
| 284 |
+
TCGA-CV-7104-01,1,12.92040635
|
| 285 |
+
TCGA-CV-7177-01,1,21.7970214
|
| 286 |
+
TCGA-CV-7178-01,1,71.21017852
|
| 287 |
+
TCGA-CV-7180-01,1,10.75056712
|
| 288 |
+
TCGA-CV-7183-01,0,130.8807575
|
| 289 |
+
TCGA-CV-7235-01,0,77.16079824
|
| 290 |
+
TCGA-CV-7236-01,1,4.734194694
|
| 291 |
+
TCGA-CV-7238-01,0,89.65381201
|
| 292 |
+
TCGA-CV-7242-01,0,35.99960548
|
| 293 |
+
TCGA-CV-7243-01,0,31.36403985
|
| 294 |
+
TCGA-CV-7245-01,0,26.20245258
|
| 295 |
+
TCGA-CV-7247-01,1,18.96965513
|
| 296 |
+
TCGA-CV-7248-01,1,17.12857941
|
| 297 |
+
TCGA-CV-7250-01,1,95.34142092
|
| 298 |
+
TCGA-CV-7252-01,1,4.964329158
|
| 299 |
+
TCGA-CV-7253-01,1,11.86836309
|
| 300 |
+
TCGA-CV-7254-01,1,47.96659763
|
| 301 |
+
TCGA-CV-7255-01,1,2.104086531
|
| 302 |
+
TCGA-CV-7261-01,0,49.70904428
|
| 303 |
+
TCGA-CV-7263-01,1,18.41075714
|
| 304 |
+
TCGA-CV-7406-01,1,57.46786337
|
| 305 |
+
TCGA-CV-7407-01,1,35.53933656
|
| 306 |
+
TCGA-CV-7409-01,1,17.85185916
|
| 307 |
+
TCGA-CV-7410-01,1,210.967551
|
| 308 |
+
TCGA-CV-7411-01,1,89.32504849
|
| 309 |
+
TCGA-CV-7413-01,1,9.6656475
|
| 310 |
+
TCGA-CV-7414-01,1,0.460268929
|
| 311 |
+
TCGA-CV-7415-01,1,22.84906467
|
| 312 |
+
TCGA-CV-7416-01,1,25.08465661
|
| 313 |
+
TCGA-CV-7418-01,1,25.93944176
|
| 314 |
+
TCGA-CV-7421-01,1,0.065752704
|
| 315 |
+
TCGA-CV-7422-01,1,34.09277707
|
| 316 |
+
TCGA-CV-7423-01,1,100.5687609
|
| 317 |
+
TCGA-CV-7424-01,1,14.89298747
|
| 318 |
+
TCGA-CV-7425-01,1,56.48157281
|
| 319 |
+
TCGA-CV-7427-01,1,156.4914357
|
| 320 |
+
TCGA-CV-7428-01,1,54.93638426
|
| 321 |
+
TCGA-CV-7429-01,1,3.517769668
|
| 322 |
+
TCGA-CV-7430-01,1,16.27379426
|
| 323 |
+
TCGA-CV-7432-01,1,84.49222474
|
| 324 |
+
TCGA-CV-7433-01,1,19.75868758
|
| 325 |
+
TCGA-CV-7434-01,1,7.167044745
|
| 326 |
+
TCGA-CV-7435-01,1,153.8613276
|
| 327 |
+
TCGA-CV-7437-01,1,16.63543413
|
| 328 |
+
TCGA-CV-7438-01,1,6.378012296
|
| 329 |
+
TCGA-CV-7440-01,1,22.19153763
|
| 330 |
+
TCGA-CV-7446-01,1,35.93385278
|
| 331 |
+
TCGA-CV-7568-01,1,30.47637834
|
| 332 |
+
TCGA-CV-A45O-01,0,27.97777559
|
| 333 |
+
TCGA-CV-A45P-01,0,21.00798895
|
| 334 |
+
TCGA-CV-A45Q-01,1,169.3789657
|
| 335 |
+
TCGA-CV-A45R-01,0,180.1624092
|
| 336 |
+
TCGA-CV-A45T-01,1,159.6475655
|
| 337 |
+
TCGA-CV-A45U-01,1,35.47358385
|
| 338 |
+
TCGA-CV-A45V-01,1,1.052043265
|
| 339 |
+
TCGA-CV-A45W-01,1,45.96114015
|
| 340 |
+
TCGA-CV-A45X-01,1,6.509517704
|
| 341 |
+
TCGA-CV-A45Y-01,1,88.86477956
|
| 342 |
+
TCGA-CV-A45Z-01,1,48.19673209
|
| 343 |
+
TCGA-CV-A460-01,1,60.42673505
|
| 344 |
+
TCGA-CV-A461-01,1,67.85679061
|
| 345 |
+
TCGA-CV-A463-01,1,0.756156097
|
| 346 |
+
TCGA-CV-A464-01,0,56.61307821
|
| 347 |
+
TCGA-CV-A465-01,1,7.068415689
|
| 348 |
+
TCGA-CV-A468-01,1,15.25462735
|
| 349 |
+
TCGA-CV-A6JD-01,1,5.983496071
|
| 350 |
+
TCGA-CV-A6JE-01,0,35.34207844
|
| 351 |
+
TCGA-CV-A6JM-01,1,6.378012296
|
| 352 |
+
TCGA-CV-A6JN-01,0,29.78597495
|
| 353 |
+
TCGA-CV-A6JO-01,1,6.476641352
|
| 354 |
+
TCGA-CV-A6JT-01,0,28.01065194
|
| 355 |
+
TCGA-CV-A6JU-01,0,3.616398724
|
| 356 |
+
TCGA-CV-A6JY-01,0,21.23812342
|
| 357 |
+
TCGA-CV-A6JZ-01,0,23.47371536
|
| 358 |
+
TCGA-CV-A6K0-01,0,19.92306934
|
| 359 |
+
TCGA-CV-A6K1-01,0,22.52030115
|
| 360 |
+
TCGA-CV-A6K2-01,1,10.4218036
|
| 361 |
+
TCGA-CX-7085-01,0,10.55330901
|
| 362 |
+
TCGA-CX-7086-01,0,18.83814972
|
| 363 |
+
TCGA-CX-7219-01,0,34.35578788
|
| 364 |
+
TCGA-CX-A4AQ-01,0,51.12272742
|
| 365 |
+
TCGA-D6-6515-01,1,13.24916987
|
| 366 |
+
TCGA-D6-6516-01,0,25.41342013
|
| 367 |
+
TCGA-D6-6517-01,0,9.599894796
|
| 368 |
+
TCGA-D6-6823-01,0,23.04632278
|
| 369 |
+
TCGA-D6-6824-01,0,2.531479107
|
| 370 |
+
TCGA-D6-6825-01,0,16.14228885
|
| 371 |
+
TCGA-D6-6826-01,1,11.44097051
|
| 372 |
+
TCGA-D6-6827-01,0,18.67376796
|
| 373 |
+
TCGA-D6-8568-01,0,24.9531512
|
| 374 |
+
TCGA-D6-8569-01,0,25.31479107
|
| 375 |
+
TCGA-D6-A4Z9-01,0,17.72035375
|
| 376 |
+
TCGA-D6-A4ZB-01,0,12.36150837
|
| 377 |
+
TCGA-D6-A6EK-01,0,28.76680804
|
| 378 |
+
TCGA-D6-A6EM-01,0,7.627313673
|
| 379 |
+
TCGA-D6-A6EN-01,0,22.58605385
|
| 380 |
+
TCGA-D6-A6EO-01,0,24.9531512
|
| 381 |
+
TCGA-D6-A6EP-01,0,13.93957327
|
| 382 |
+
TCGA-D6-A6EQ-01,0,12.09849755
|
| 383 |
+
TCGA-D6-A6ES-01,0,12.78890094
|
| 384 |
+
TCGA-D6-A74Q-01,0,23.34220995
|
| 385 |
+
TCGA-DQ-5624-01,0,58.45415393
|
| 386 |
+
TCGA-DQ-5625-01,1,37.24890686
|
| 387 |
+
TCGA-DQ-5629-01,1,30.93664727
|
| 388 |
+
TCGA-DQ-5630-01,0,33.8626426
|
| 389 |
+
TCGA-DQ-5631-01,1,18.01624092
|
| 390 |
+
TCGA-DQ-7588-01,1,14.03820232
|
| 391 |
+
TCGA-DQ-7589-01,0,46.32278002
|
| 392 |
+
TCGA-DQ-7590-01,0,46.45428543
|
| 393 |
+
TCGA-DQ-7591-01,0,20.44909097
|
| 394 |
+
TCGA-DQ-7592-01,0,37.57767038
|
| 395 |
+
TCGA-DQ-7593-01,0,40.2406549
|
| 396 |
+
TCGA-DQ-7594-01,0,40.04339679
|
| 397 |
+
TCGA-DQ-7595-01,0,39.12285893
|
| 398 |
+
TCGA-DQ-7596-01,0,41.58858533
|
| 399 |
+
TCGA-F7-7848-01,0,37.18315416
|
| 400 |
+
TCGA-F7-8298-01,0,32.71197028
|
| 401 |
+
TCGA-F7-8489-01,0,21.63263964
|
| 402 |
+
TCGA-F7-A50G-01,0,20.25183286
|
| 403 |
+
TCGA-F7-A50I-01,0,3.024624388
|
| 404 |
+
TCGA-F7-A50J-01,0,31.13390538
|
| 405 |
+
TCGA-F7-A61S-01,0,18.93677878
|
| 406 |
+
TCGA-F7-A61V-01,0,24.9531512
|
| 407 |
+
TCGA-F7-A61W-01,0,0.460268929
|
| 408 |
+
TCGA-F7-A620-01,0,17.85185916
|
| 409 |
+
TCGA-F7-A622-01,1,11.80261038
|
| 410 |
+
TCGA-F7-A623-01,0,20.25183286
|
| 411 |
+
TCGA-F7-A624-01,0,12.42726107
|
| 412 |
+
TCGA-H7-7774-01,0,13.38067528
|
| 413 |
+
TCGA-H7-8501-01,0,15.15599829
|
| 414 |
+
TCGA-H7-8502-01,0,15.05736923
|
| 415 |
+
TCGA-H7-A6C4-01,0,13.61080975
|
| 416 |
+
TCGA-H7-A6C5-01,0,21.10661801
|
| 417 |
+
TCGA-H7-A76A-01,0,20.94223625
|
| 418 |
+
TCGA-HD-7229-01,0,33.76401355
|
| 419 |
+
TCGA-HD-7753-01,0,28.47092087
|
| 420 |
+
TCGA-HD-7754-01,0,25.74218365
|
| 421 |
+
TCGA-HD-7831-01,0,21.92852681
|
| 422 |
+
TCGA-HD-7832-01,0,27.48463031
|
| 423 |
+
TCGA-HD-7917-01,1,27.48463031
|
| 424 |
+
TCGA-HD-8224-01,1,14.66285301
|
| 425 |
+
TCGA-HD-8314-01,0,22.02715587
|
| 426 |
+
TCGA-HD-8634-01,1,12.65739554
|
| 427 |
+
TCGA-HD-8635-01,0,22.84906467
|
| 428 |
+
TCGA-HD-A4C1-01,0,0.361639872
|
| 429 |
+
TCGA-HD-A633-01,0,13.84094421
|
| 430 |
+
TCGA-HD-A634-01,1,4.273925765
|
| 431 |
+
TCGA-HD-A6HZ-01,0,3.649275076
|
| 432 |
+
TCGA-HD-A6I0-01,0,6.904033928
|
| 433 |
+
TCGA-HL-7533-01,0,34.75030411
|
| 434 |
+
TCGA-IQ-7630-01,0,15.94503074
|
| 435 |
+
TCGA-IQ-7631-01,0,38.53108459
|
| 436 |
+
TCGA-IQ-7632-01,0,14.49847125
|
| 437 |
+
TCGA-IQ-A61E-01,0,37.70917579
|
| 438 |
+
TCGA-IQ-A61G-01,0,11.83548673
|
| 439 |
+
TCGA-IQ-A61H-01,0,37.41328862
|
| 440 |
+
TCGA-IQ-A61I-01,1,0.065752704
|
| 441 |
+
TCGA-IQ-A61J-01,0,33.56675543
|
| 442 |
+
TCGA-IQ-A61K-01,1,5.293092678
|
| 443 |
+
TCGA-IQ-A61L-01,0,13.67656245
|
| 444 |
+
TCGA-IQ-A61O-01,1,13.84094421
|
| 445 |
+
TCGA-IQ-A6SG-01,0,19.03540783
|
| 446 |
+
TCGA-IQ-A6SH-01,0,15.48476181
|
| 447 |
+
TCGA-KU-A66S-01,1,13.34779893
|
| 448 |
+
TCGA-KU-A66T-01,0,18.14774633
|
| 449 |
+
TCGA-KU-A6H7-01,0,19.2655423
|
| 450 |
+
TCGA-KU-A6H8-01,1,10.75056712
|
| 451 |
+
TCGA-MT-A51W-01,0,14.36696584
|
| 452 |
+
TCGA-MT-A51X-01,0,7.956077194
|
| 453 |
+
TCGA-MT-A67A-01,0,30.04898577
|
| 454 |
+
TCGA-MT-A67D-01,0,1.841075714
|
| 455 |
+
TCGA-MT-A67F-01,0,12.62451918
|
| 456 |
+
TCGA-MT-A67G-01,0,6.246506888
|
| 457 |
+
TCGA-MT-A7BN-01,0,15.41900911
|
| 458 |
+
TCGA-MZ-A5BI-01,1,7.134168393
|
| 459 |
+
TCGA-MZ-A6I9-01,1,16.07653615
|
| 460 |
+
TCGA-MZ-A7D7-01,0,17.98336457
|
| 461 |
+
TCGA-P3-A5Q5-01,0,29.91748036
|
| 462 |
+
TCGA-P3-A5Q6-01,1,15.78064898
|
| 463 |
+
TCGA-P3-A5QA-01,0,71.73620015
|
| 464 |
+
TCGA-P3-A5QE-01,0,51.25423283
|
| 465 |
+
TCGA-P3-A5QF-01,1,10.84919617
|
| 466 |
+
TCGA-P3-A6SW-01,0,36.82151429
|
| 467 |
+
TCGA-P3-A6SX-01,1,47.01318342
|
| 468 |
+
TCGA-P3-A6T0-01,0,19.00253148
|
| 469 |
+
TCGA-P3-A6T2-01,0,75.54985699
|
| 470 |
+
TCGA-P3-A6T3-01,1,18.96965513
|
| 471 |
+
TCGA-P3-A6T4-01,1,2.038333827
|
| 472 |
+
TCGA-P3-A6T5-01,1,28.9969425
|
| 473 |
+
TCGA-P3-A6T6-01,1,12.98615906
|
| 474 |
+
TCGA-P3-A6T7-01,1,16.01078344
|
| 475 |
+
TCGA-P3-A6T8-01,0,13.15054082
|
| 476 |
+
TCGA-QK-A64Z-01,1,21.07374166
|
| 477 |
+
TCGA-QK-A652-01,0,21.20524707
|
| 478 |
+
TCGA-QK-A6IF-01,0,23.14495184
|
| 479 |
+
TCGA-QK-A6IG-01,1,7.298550153
|
| 480 |
+
TCGA-QK-A6IH-01,0,21.46825788
|
| 481 |
+
TCGA-QK-A6II-01,1,9.336883979
|
| 482 |
+
TCGA-QK-A6IJ-01,0,12.72314824
|
| 483 |
+
TCGA-QK-A6V9-01,0,27.38600125
|
| 484 |
+
TCGA-QK-A6VB-01,0,21.07374166
|
| 485 |
+
TCGA-QK-A6VC-01,0,19.72581122
|
| 486 |
+
TCGA-QK-A8Z7-01,0,12.88753
|
| 487 |
+
TCGA-QK-A8Z8-01,1,5.621856199
|
| 488 |
+
TCGA-QK-A8Z9-01,1,14.76148207
|
| 489 |
+
TCGA-QK-A8ZA-01,1,12.19712661
|
| 490 |
+
TCGA-QK-A8ZB-01,0,17.81898281
|
| 491 |
+
TCGA-QK-AA3J-01,0,15.32038005
|
| 492 |
+
TCGA-QK-AA3K-01,0,8.317717066
|
| 493 |
+
TCGA-RS-A6TO-01,1,12.72314824
|
| 494 |
+
TCGA-RS-A6TP-01,0,16.96419765
|
| 495 |
+
TCGA-T2-A6WX-01,1,6.871157576
|
| 496 |
+
TCGA-T2-A6WZ-01,1,15.91215439
|
| 497 |
+
TCGA-T2-A6X0-01,0,7.101292041
|
| 498 |
+
TCGA-T2-A6X2-01,0,32.44895946
|
| 499 |
+
TCGA-T3-A92M-01,0,13.7094388
|
| 500 |
+
TCGA-T3-A92N-01,1,3.123253444
|
| 501 |
+
TCGA-TN-A7HI-01,0,13.54505704
|
| 502 |
+
TCGA-TN-A7HJ-01,0,13.24916987
|
| 503 |
+
TCGA-TN-A7HL-01,0,20.35046191
|
| 504 |
+
TCGA-UF-A718-01,0,64.79928987
|
| 505 |
+
TCGA-UF-A719-01,0,54.67337344
|
| 506 |
+
TCGA-UF-A71A-01,1,2.827366275
|
| 507 |
+
TCGA-UF-A71B-01,0,49.51178617
|
| 508 |
+
TCGA-UF-A71D-01,0,48.03235033
|
| 509 |
+
TCGA-UF-A71E-01,1,49.44603347
|
| 510 |
+
TCGA-UF-A7J9-01,0,44.64608607
|
| 511 |
+
TCGA-UF-A7JA-01,0,74.46493737
|
| 512 |
+
TCGA-UF-A7JC-01,1,17.95048821
|
| 513 |
+
TCGA-UF-A7JD-01,1,24.29562416
|
| 514 |
+
TCGA-UF-A7JF-01,0,55.42952954
|
| 515 |
+
TCGA-UF-A7JH-01,0,29.45721143
|
| 516 |
+
TCGA-UF-A7JJ-01,0,18.04911727
|
| 517 |
+
TCGA-UF-A7JK-01,1,13.93957327
|
| 518 |
+
TCGA-UF-A7JO-01,1,20.74497814
|
| 519 |
+
TCGA-UF-A7JS-01,1,22.35591939
|
| 520 |
+
TCGA-UF-A7JT-01,1,32.64621758
|
| 521 |
+
TCGA-UF-A7JV-01,1,2.958871684
|
| 522 |
+
TCGA-UP-A6WW-01,0,17.02995036
|
| 523 |
+
TCGA-WA-A7GZ-01,1,20.54772003
|
| 524 |
+
TCGA-WA-A7H4-01,0,14.56422395
|
lc_models/MultiOmicsAutoencoder/trained_autoencoder/config.json
ADDED
|
@@ -0,0 +1,22 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"architectures": [
|
| 3 |
+
"MultiOmicsGraphAttentionAutoencoderModel"
|
| 4 |
+
],
|
| 5 |
+
"edge_attr_channels": 1,
|
| 6 |
+
"edge_decoder_activations": [
|
| 7 |
+
"ReLU",
|
| 8 |
+
"ReLU"
|
| 9 |
+
],
|
| 10 |
+
"edge_decoder_hidden_sizes": [
|
| 11 |
+
128,
|
| 12 |
+
64
|
| 13 |
+
],
|
| 14 |
+
"in_channels": 17,
|
| 15 |
+
"learning_rate": 0.01,
|
| 16 |
+
"model_type": "omics-graph-network",
|
| 17 |
+
"num_layers": 2,
|
| 18 |
+
"original_feature_size": 17,
|
| 19 |
+
"out_channels": 1,
|
| 20 |
+
"torch_dtype": "float32",
|
| 21 |
+
"transformers_version": "4.32.1"
|
| 22 |
+
}
|
lc_models/MultiOmicsAutoencoder/trained_autoencoder/pytorch_model.bin
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:9651f8c59ac5c0c57494a6553c92a14d78427ea6bbd0e2474132b1eea6b4ec87
|
| 3 |
+
size 43263
|
lc_models/MultiOmicsAutoencoder/trained_decoder/config.json
ADDED
|
@@ -0,0 +1,22 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"architectures": [
|
| 3 |
+
"GATv2DecoderModel"
|
| 4 |
+
],
|
| 5 |
+
"edge_attr_channels": 1,
|
| 6 |
+
"edge_decoder_activations": [
|
| 7 |
+
"ReLU",
|
| 8 |
+
"ReLU"
|
| 9 |
+
],
|
| 10 |
+
"edge_decoder_hidden_sizes": [
|
| 11 |
+
128,
|
| 12 |
+
64
|
| 13 |
+
],
|
| 14 |
+
"in_channels": 17,
|
| 15 |
+
"learning_rate": 0.01,
|
| 16 |
+
"model_type": "omics-graph-network",
|
| 17 |
+
"num_layers": 2,
|
| 18 |
+
"original_feature_size": 17,
|
| 19 |
+
"out_channels": 1,
|
| 20 |
+
"torch_dtype": "float32",
|
| 21 |
+
"transformers_version": "4.32.1"
|
| 22 |
+
}
|
lc_models/MultiOmicsAutoencoder/trained_decoder/pytorch_model.bin
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:3c49a50b01c9f04d3ec0cfd756e14d982e619911b86e1bb96d056dee541982f5
|
| 3 |
+
size 38933
|
lc_models/MultiOmicsAutoencoder/trained_edge_weight_predictor/config.json
ADDED
|
@@ -0,0 +1,22 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"architectures": [
|
| 3 |
+
"EdgeWeightPredictorModel"
|
| 4 |
+
],
|
| 5 |
+
"edge_attr_channels": 1,
|
| 6 |
+
"edge_decoder_activations": [
|
| 7 |
+
"ReLU",
|
| 8 |
+
"ReLU"
|
| 9 |
+
],
|
| 10 |
+
"edge_decoder_hidden_sizes": [
|
| 11 |
+
128,
|
| 12 |
+
64
|
| 13 |
+
],
|
| 14 |
+
"in_channels": 17,
|
| 15 |
+
"learning_rate": 0.01,
|
| 16 |
+
"model_type": "omics-graph-network",
|
| 17 |
+
"num_layers": 2,
|
| 18 |
+
"original_feature_size": 17,
|
| 19 |
+
"out_channels": 1,
|
| 20 |
+
"torch_dtype": "float32",
|
| 21 |
+
"transformers_version": "4.32.1"
|
| 22 |
+
}
|
lc_models/MultiOmicsAutoencoder/trained_edge_weight_predictor/pytorch_model.bin
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:93767ffe12d84ff9af955d7704c638317aafa046c58ad06e09a846513fd18f1b
|
| 3 |
+
size 36999
|
lc_models/MultiOmicsAutoencoder/trained_encoder/config.json
ADDED
|
@@ -0,0 +1,22 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"architectures": [
|
| 3 |
+
"GATv2EncoderModel"
|
| 4 |
+
],
|
| 5 |
+
"edge_attr_channels": 1,
|
| 6 |
+
"edge_decoder_activations": [
|
| 7 |
+
"ReLU",
|
| 8 |
+
"ReLU"
|
| 9 |
+
],
|
| 10 |
+
"edge_decoder_hidden_sizes": [
|
| 11 |
+
128,
|
| 12 |
+
64
|
| 13 |
+
],
|
| 14 |
+
"in_channels": 17,
|
| 15 |
+
"learning_rate": 0.01,
|
| 16 |
+
"model_type": "omics-graph-network",
|
| 17 |
+
"num_layers": 2,
|
| 18 |
+
"original_feature_size": 17,
|
| 19 |
+
"out_channels": 1,
|
| 20 |
+
"torch_dtype": "float32",
|
| 21 |
+
"transformers_version": "4.32.1"
|
| 22 |
+
}
|
lc_models/MultiOmicsAutoencoder/trained_encoder/pytorch_model.bin
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:73804072e5ca45ce24a20496c86ee124569589a5efda620aa589843a5ad83966
|
| 3 |
+
size 4571
|
results/temp_k3_club_plan_meir.jpeg
ADDED
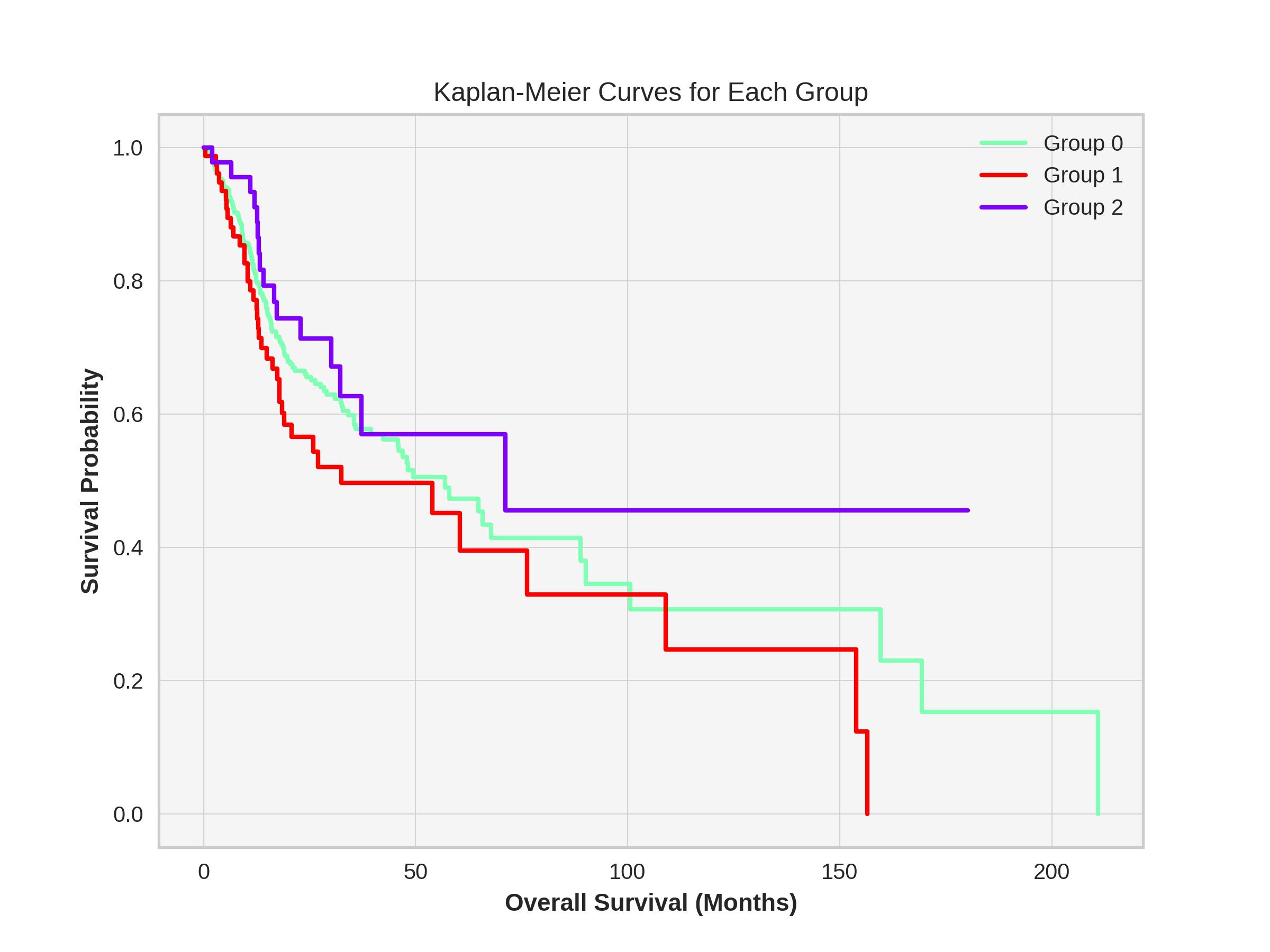
|
results/temp_k3_club_plan_meir.png
ADDED

|
results/temp_k5_plan_meir.jpeg
ADDED

|
results/temp_k5_plan_meir.png
ADDED
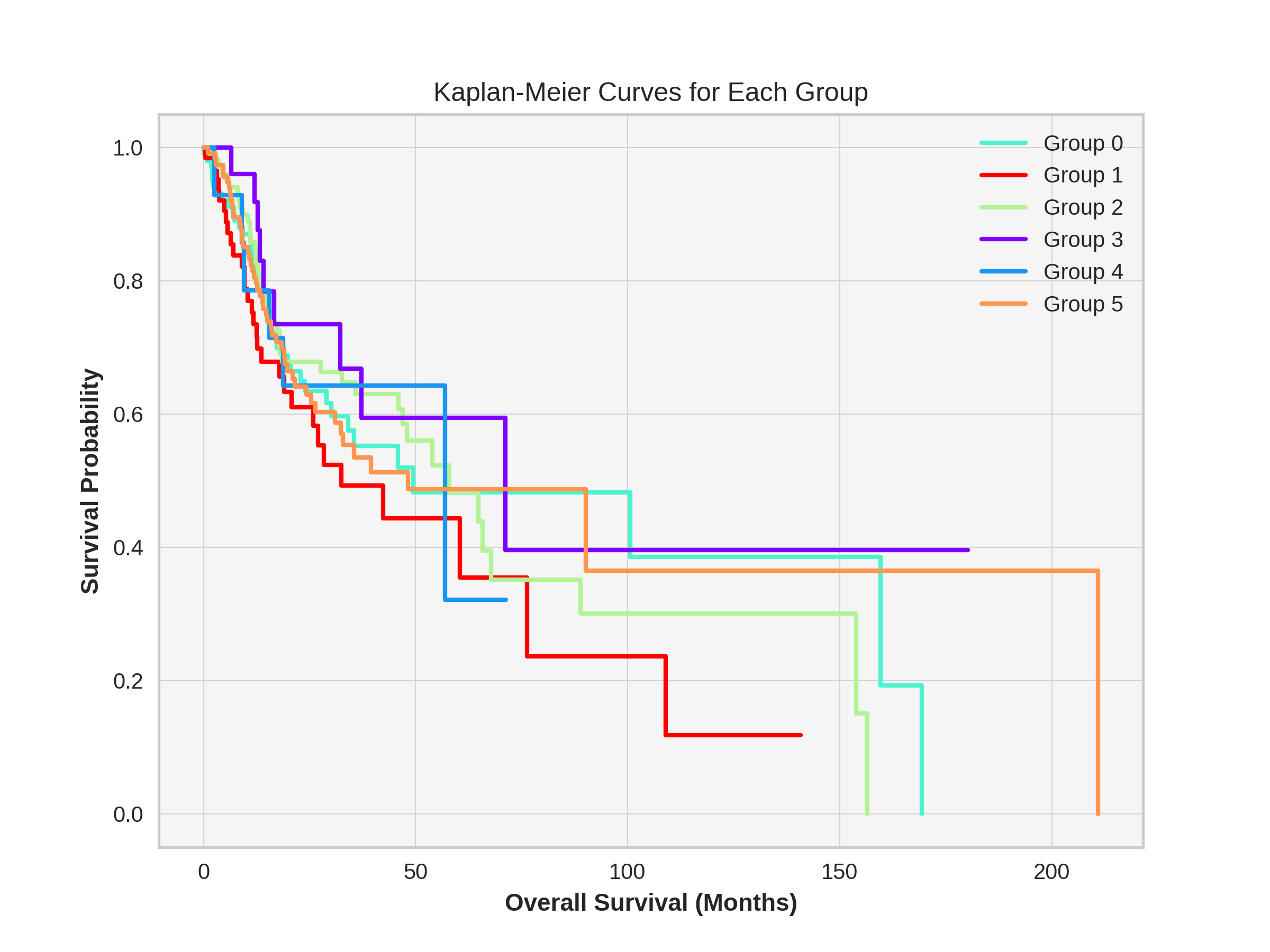
|
results/temp_median_survival.jpeg
ADDED

|
results/temp_median_survival.png
ADDED

|
train.py
ADDED
|
@@ -0,0 +1,105 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import torch
|
| 2 |
+
from torch.utils.data import DataLoader
|
| 3 |
+
from torch_geometric.data import Batch
|
| 4 |
+
from sklearn.model_selection import train_test_split
|
| 5 |
+
import pickle
|
| 6 |
+
|
| 7 |
+
from OmicsConfig import OmicsConfig
|
| 8 |
+
from MultiOmicsGraphAttentionAutoencoderModel import MultiOmicsGraphAttentionAutoencoderModel
|
| 9 |
+
from GATv2EncoderModel import GATv2EncoderModel
|
| 10 |
+
from GATv2DecoderModel import GATv2DecoderModel
|
| 11 |
+
from EdgeWeightPredictorModel import EdgeWeightPredictorModel
|
| 12 |
+
|
| 13 |
+
def collate_graph_data(batch):
|
| 14 |
+
return Batch.from_data_list(batch)
|
| 15 |
+
|
| 16 |
+
def create_data_loader(graph_data_dict, batch_size=1, shuffle=True):
|
| 17 |
+
graph_data = list(graph_data_dict.values())
|
| 18 |
+
return DataLoader(graph_data, batch_size=batch_size, shuffle=shuffle, collate_fn=collate_graph_data)
|
| 19 |
+
|
| 20 |
+
# Load your data
|
| 21 |
+
graph_data_dict = torch.load('data/graph_data_dictN.pth')
|
| 22 |
+
|
| 23 |
+
# Split the data
|
| 24 |
+
train_data, temp_data = train_test_split(list(graph_data_dict.items()), train_size=0.6, random_state=42)
|
| 25 |
+
val_data, test_data = train_test_split(temp_data, test_size=0.5, random_state=42)
|
| 26 |
+
|
| 27 |
+
# Convert lists back into dictionaries
|
| 28 |
+
train_data = dict(train_data)
|
| 29 |
+
val_data = dict(val_data)
|
| 30 |
+
test_data = dict(test_data)
|
| 31 |
+
|
| 32 |
+
# Define the configuration for the model
|
| 33 |
+
autoencoder_config = OmicsConfig(
|
| 34 |
+
in_channels=17,
|
| 35 |
+
edge_attr_channels=1,
|
| 36 |
+
out_channels=1,
|
| 37 |
+
original_feature_size=17,
|
| 38 |
+
learning_rate=0.01,
|
| 39 |
+
num_layers=2,
|
| 40 |
+
edge_decoder_hidden_sizes=[128, 64],
|
| 41 |
+
edge_decoder_activations=['ReLU', 'ReLU']
|
| 42 |
+
)
|
| 43 |
+
|
| 44 |
+
# Initialize the model
|
| 45 |
+
autoencoder_model = MultiOmicsGraphAttentionAutoencoderModel(autoencoder_config)
|
| 46 |
+
|
| 47 |
+
# Create data loaders
|
| 48 |
+
train_loader = create_data_loader(train_data, batch_size=4, shuffle=True)
|
| 49 |
+
val_loader = create_data_loader(val_data, batch_size=4, shuffle=False)
|
| 50 |
+
test_loader = create_data_loader(test_data, batch_size=4, shuffle=False)
|
| 51 |
+
|
| 52 |
+
# Define the device
|
| 53 |
+
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
|
| 54 |
+
|
| 55 |
+
# Training process
|
| 56 |
+
def train_autoencoder(autoencoder_model, train_loader, validation_loader, epochs, device):
|
| 57 |
+
autoencoder_model.to(device)
|
| 58 |
+
train_losses = []
|
| 59 |
+
val_losses = []
|
| 60 |
+
|
| 61 |
+
for epoch in range(epochs):
|
| 62 |
+
# Train
|
| 63 |
+
autoencoder_model.train()
|
| 64 |
+
train_loss, train_cosine_similarity = autoencoder_model.train_model(train_loader, device)
|
| 65 |
+
print(f"Epoch {epoch+1}/{epochs}, Train Loss: {train_loss:.4f}, Train Cosine Similarity: {train_cosine_similarity:.4f}")
|
| 66 |
+
train_losses.append(train_loss)
|
| 67 |
+
|
| 68 |
+
# Validate
|
| 69 |
+
autoencoder_model.eval()
|
| 70 |
+
val_loss, val_cosine_similarity = autoencoder_model.validate(validation_loader, device)
|
| 71 |
+
print(f"Epoch {epoch+1}/{epochs}, Validation Loss: {val_loss:.4f}, Validation Cosine Similarity: {val_cosine_similarity:.4f}")
|
| 72 |
+
val_losses.append(val_loss)
|
| 73 |
+
|
| 74 |
+
# Save the trained encoder weights
|
| 75 |
+
trained_encoder_path = "lc_models/MultiOmicsAutoencoder/trained_encoder"
|
| 76 |
+
autoencoder_model.encoder.save_pretrained(trained_encoder_path)
|
| 77 |
+
|
| 78 |
+
# Save the trained decoder weights
|
| 79 |
+
trained_decoder_path = "lc_models/MultiOmicsAutoencoder/trained_decoder"
|
| 80 |
+
autoencoder_model.decoder.save_pretrained(trained_decoder_path)
|
| 81 |
+
|
| 82 |
+
# Save the trained edge weight predictor weights (if needed separately)
|
| 83 |
+
trained_edge_weight_predictor_path = "lc_models/MultiOmicsAutoencoder/trained_edge_weight_predictor"
|
| 84 |
+
autoencoder_model.decoder.edge_weight_predictor.save_pretrained(trained_edge_weight_predictor_path)
|
| 85 |
+
|
| 86 |
+
# Optionally save the entire autoencoder again if you want to have a complete package
|
| 87 |
+
trained_autoencoder_path = "lc_models/MultiOmicsAutoencoder/trained_autoencoder"
|
| 88 |
+
autoencoder_model.save_pretrained(trained_autoencoder_path)
|
| 89 |
+
|
| 90 |
+
return train_losses, val_losses
|
| 91 |
+
|
| 92 |
+
# Train and save the model
|
| 93 |
+
train_losses, val_losses = train_autoencoder(autoencoder_model, train_loader, val_loader, epochs=10, device=device)
|
| 94 |
+
|
| 95 |
+
# Evaluate the model
|
| 96 |
+
test_loss, test_accuracy = autoencoder_model.evaluate(test_loader, device)
|
| 97 |
+
print(f"Test Loss: {test_loss:.4f}")
|
| 98 |
+
print(f"Test Accuracy: {test_accuracy:.4%}")
|
| 99 |
+
|
| 100 |
+
# Save the training and validation losses
|
| 101 |
+
with open('./results/train_loss.pkl', 'wb') as f:
|
| 102 |
+
pickle.dump(train_losses, f)
|
| 103 |
+
|
| 104 |
+
with open('./results/val_loss.pkl', 'wb') as f:
|
| 105 |
+
pickle.dump(val_losses, f)
|