Spaces:
Runtime error
Runtime error
Upload folder using huggingface_hub
Browse files- .dockerignore +2 -0
- .zen/config.yaml +2 -0
- Dockerfile +26 -0
- README.md +53 -12
- _assets/airflow_stack.png +0 -0
- _assets/default_stack.png +0 -0
- _assets/local_sagmaker_so_stack.png +0 -0
- _assets/sagemaker_stack.png +0 -0
- app.py +48 -0
- configs/deployment.yaml +13 -0
- configs/feature_engineering.yaml +12 -0
- configs/inference.yaml +13 -0
- configs/training.yaml +12 -0
- flagged/log.csv +2 -0
- flagged/output/tmpjy2eamkw.json +1 -0
- pipelines/__init__.py +6 -0
- pipelines/__pycache__/__init__.cpython-38.pyc +0 -0
- pipelines/__pycache__/deployment.cpython-38.pyc +0 -0
- pipelines/__pycache__/feature_engineering.cpython-38.pyc +0 -0
- pipelines/__pycache__/inference.cpython-38.pyc +0 -0
- pipelines/__pycache__/training.cpython-38.pyc +0 -0
- pipelines/deployment.py +38 -0
- pipelines/feature_engineering.py +54 -0
- pipelines/inference.py +50 -0
- pipelines/training.py +61 -0
- requirements.txt +3 -0
- run.ipynb +981 -0
- run.py +173 -0
- run_stack_showcase.ipynb +347 -0
- steps/__init__.py +29 -0
- steps/__pycache__/__init__.cpython-38.pyc +0 -0
- steps/__pycache__/data_loader.cpython-38.pyc +0 -0
- steps/__pycache__/data_preprocessor.cpython-38.pyc +0 -0
- steps/__pycache__/data_splitter.cpython-38.pyc +0 -0
- steps/__pycache__/deploy_to_huggingface.cpython-38.pyc +0 -0
- steps/__pycache__/inference_predict.cpython-38.pyc +0 -0
- steps/__pycache__/inference_preprocessor.cpython-38.pyc +0 -0
- steps/__pycache__/model_evaluator.cpython-38.pyc +0 -0
- steps/__pycache__/model_promoter.cpython-38.pyc +0 -0
- steps/__pycache__/model_trainer.cpython-38.pyc +0 -0
- steps/data_loader.py +53 -0
- steps/data_preprocessor.py +115 -0
- steps/data_splitter.py +47 -0
- steps/deploy_to_huggingface.py +58 -0
- steps/inference_predict.py +59 -0
- steps/inference_preprocessor.py +52 -0
- steps/model_evaluator.py +102 -0
- steps/model_promoter.py +42 -0
- steps/model_trainer.py +52 -0
.dockerignore
ADDED
|
@@ -0,0 +1,2 @@
|
|
|
|
|
|
|
|
|
|
| 1 |
+
.venv*
|
| 2 |
+
.requirements*
|
.zen/config.yaml
ADDED
|
@@ -0,0 +1,2 @@
|
|
|
|
|
|
|
|
|
|
| 1 |
+
active_stack_id: c2be0c2a-7cf0-44e7-8ee3-71400a579a27
|
| 2 |
+
active_workspace_id: f3a544f2-afb5-4672-934a-7a465c66201c
|
Dockerfile
ADDED
|
@@ -0,0 +1,26 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# read the doc: https://huggingface.co/docs/hub/spaces-sdks-docker
|
| 2 |
+
# you will also find guides on how best to write your Dockerfile
|
| 3 |
+
|
| 4 |
+
FROM python:3.9
|
| 5 |
+
|
| 6 |
+
WORKDIR /code
|
| 7 |
+
|
| 8 |
+
COPY ./requirements.txt /code/requirements.txt
|
| 9 |
+
|
| 10 |
+
RUN pip install --no-cache-dir --upgrade -r /code/requirements.txt
|
| 11 |
+
|
| 12 |
+
# Set up a new user named "user" with user ID 1000
|
| 13 |
+
RUN useradd -m -u 1000 user
|
| 14 |
+
# Switch to the "user" user
|
| 15 |
+
USER user
|
| 16 |
+
# Set home to the user's home directory
|
| 17 |
+
ENV HOME=/home/user \
|
| 18 |
+
PATH=/home/user/.local/bin:$PATH
|
| 19 |
+
|
| 20 |
+
# Set the working directory to the user's home directory
|
| 21 |
+
WORKDIR $HOME/app
|
| 22 |
+
|
| 23 |
+
# Copy the current directory contents into the container at $HOME/app setting the owner to the user
|
| 24 |
+
COPY --chown=user . $HOME/app
|
| 25 |
+
|
| 26 |
+
CMD ["python", "app.py", "--server.port=7860", "--server.address=0.0.0.0"]
|
README.md
CHANGED
|
@@ -1,12 +1,53 @@
|
|
| 1 |
-
|
| 2 |
-
|
| 3 |
-
|
| 4 |
-
|
| 5 |
-
|
| 6 |
-
|
| 7 |
-
|
| 8 |
-
|
| 9 |
-
|
| 10 |
-
|
| 11 |
-
|
| 12 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# 📜 ZenML Stack Show Case
|
| 2 |
+
|
| 3 |
+
This project aims to demonstrate the power of stacks. The code in this
|
| 4 |
+
project assumes that you have quite a few stacks registered already.
|
| 5 |
+
|
| 6 |
+
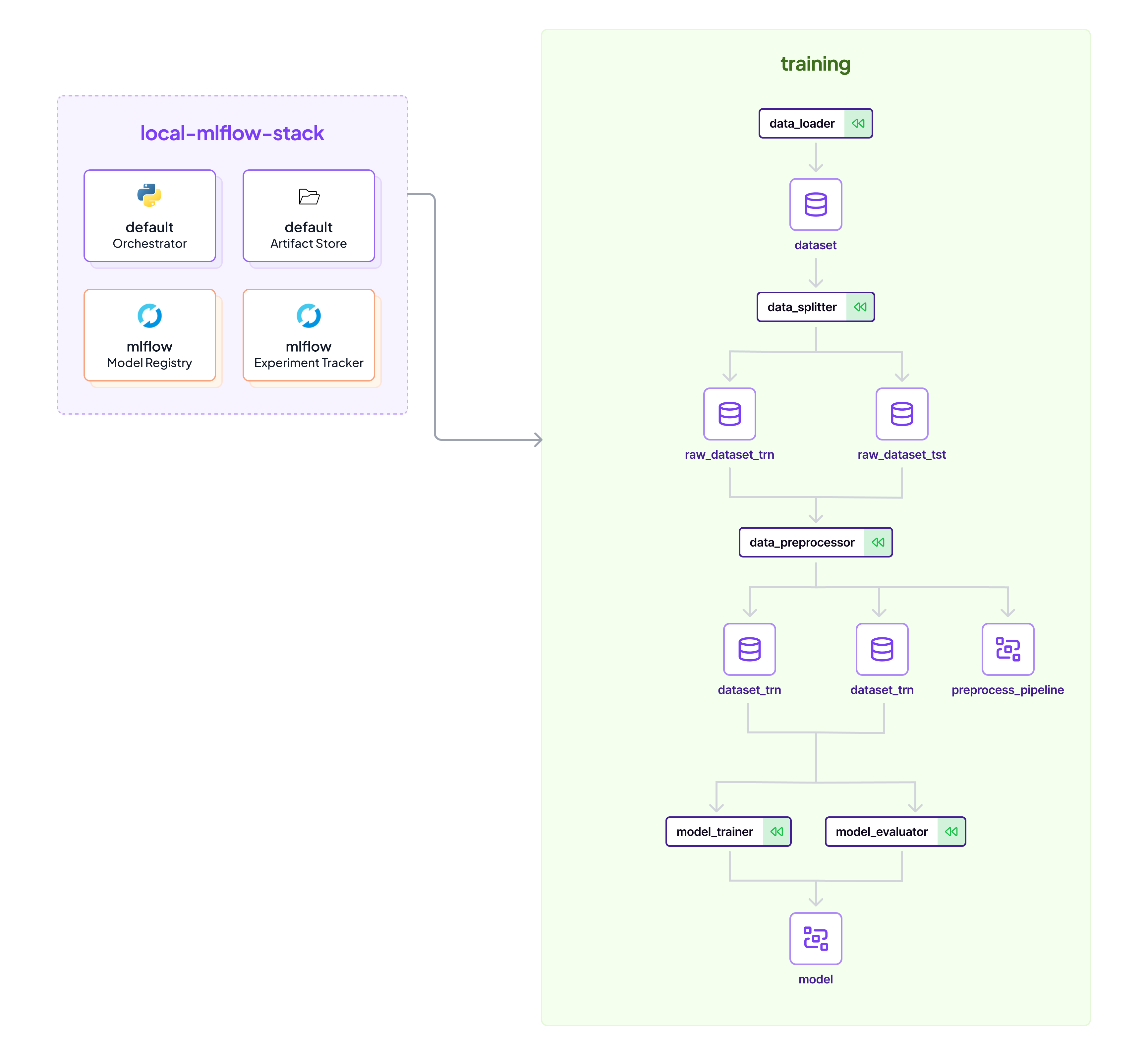
## default
|
| 7 |
+
* `default` Orchestrator
|
| 8 |
+
* `default` Artifact Store
|
| 9 |
+
|
| 10 |
+
```commandline
|
| 11 |
+
zenml stack set default
|
| 12 |
+
python run.py --training-pipeline
|
| 13 |
+
```
|
| 14 |
+
|
| 15 |
+
## local-sagemaker-step-operator-stack
|
| 16 |
+
* `default` Orchestrator
|
| 17 |
+
* `s3` Artifact Store
|
| 18 |
+
* `local` Image Builder
|
| 19 |
+
* `aws` Container Registry
|
| 20 |
+
* `Sagemaker` Step Operator
|
| 21 |
+
|
| 22 |
+
```commandline
|
| 23 |
+
zenml stack set local-sagemaker-step-operator-stack
|
| 24 |
+
zenml integration install aws -y
|
| 25 |
+
python run.py --training-pipeline
|
| 26 |
+
```
|
| 27 |
+
|
| 28 |
+
## sagemaker-airflow-stack
|
| 29 |
+
* `Airflow` Orchestrator
|
| 30 |
+
* `s3` Artifact Store
|
| 31 |
+
* `local` Image Builder
|
| 32 |
+
* `aws` Container Registry
|
| 33 |
+
* `Sagemaker` Step Operator
|
| 34 |
+
|
| 35 |
+
```commandline
|
| 36 |
+
zenml stack set sagemaker-airflow-stack
|
| 37 |
+
zenml integration install airflow -y
|
| 38 |
+
pip install apache-airflow-providers-docker apache-airflow~=2.5.0
|
| 39 |
+
zenml stack up
|
| 40 |
+
python run.py --training-pipeline
|
| 41 |
+
```
|
| 42 |
+
|
| 43 |
+
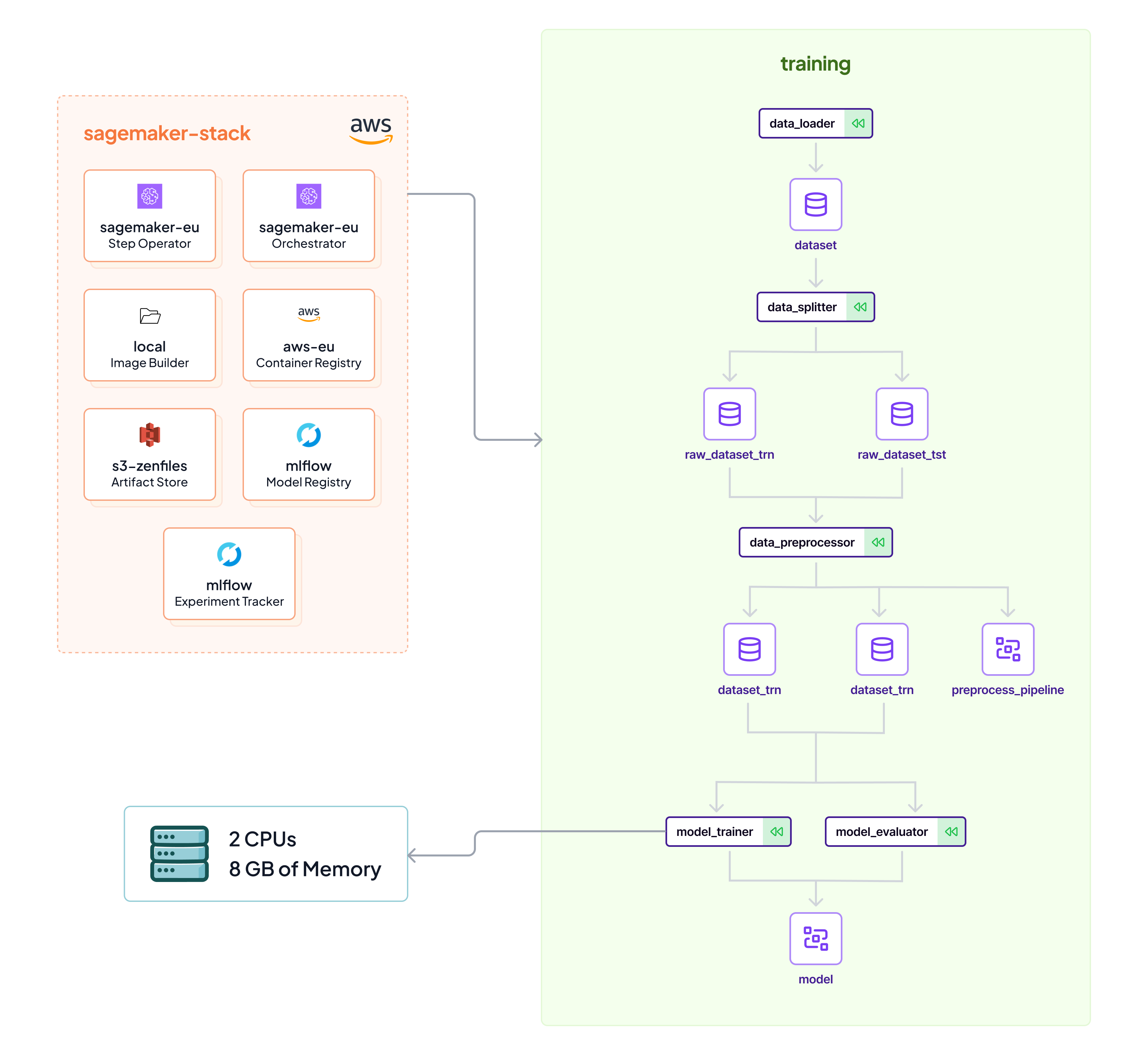
## sagemaker-stack
|
| 44 |
+
* `Sagemaker` Orchestrator
|
| 45 |
+
* `s3` Artifact Store
|
| 46 |
+
* `local` Image Builder
|
| 47 |
+
* `aws` Container Registry
|
| 48 |
+
* `Sagemaker` Step Operator
|
| 49 |
+
|
| 50 |
+
```commandline
|
| 51 |
+
zenml stack set sagemaker-stack
|
| 52 |
+
python run.py --training-pipeline
|
| 53 |
+
```
|
_assets/airflow_stack.png
ADDED

|
_assets/default_stack.png
ADDED
|
_assets/local_sagmaker_so_stack.png
ADDED

|
_assets/sagemaker_stack.png
ADDED
|
app.py
ADDED
|
@@ -0,0 +1,48 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import gradio as gr
|
| 2 |
+
import numpy as np
|
| 3 |
+
import pandas as pd
|
| 4 |
+
from sklearn.datasets import load_breast_cancer
|
| 5 |
+
from zenml.client import Client
|
| 6 |
+
|
| 7 |
+
client = Client()
|
| 8 |
+
zenml_model_version = client.get_model_version("breast_cancer_classifier", "production")
|
| 9 |
+
preprocess_pipeline = zenml_model_version.get_artifact("preprocess_pipeline").load()
|
| 10 |
+
|
| 11 |
+
# Load the model
|
| 12 |
+
clf = zenml_model_version.get_artifact("model").load()
|
| 13 |
+
|
| 14 |
+
# Load dataset to get feature names
|
| 15 |
+
data = load_breast_cancer()
|
| 16 |
+
feature_names = data.feature_names
|
| 17 |
+
|
| 18 |
+
def classify(*input_features):
|
| 19 |
+
# Convert the input features to pandas DataFrame
|
| 20 |
+
input_features = np.array(input_features).reshape(1, -1)
|
| 21 |
+
input_df = pd.DataFrame(input_features, columns=feature_names)
|
| 22 |
+
|
| 23 |
+
# Pre-process the DataFrame
|
| 24 |
+
input_df["target"] = pd.Series([1] * input_df.shape[0])
|
| 25 |
+
input_df = preprocess_pipeline.transform(input_df)
|
| 26 |
+
input_df.drop(columns=["target"], inplace=True)
|
| 27 |
+
|
| 28 |
+
# Make a prediction
|
| 29 |
+
prediction_proba = clf.predict_proba(input_df)[0]
|
| 30 |
+
|
| 31 |
+
# Map predicted class probabilities
|
| 32 |
+
classes = data.target_names
|
| 33 |
+
return {classes[idx]: prob for idx, prob in enumerate(prediction_proba)}
|
| 34 |
+
|
| 35 |
+
# Define a list of Number inputs for each feature
|
| 36 |
+
input_components = [gr.Number(label=feature_name, default=0) for feature_name in feature_names]
|
| 37 |
+
|
| 38 |
+
# Define the Gradio interface
|
| 39 |
+
iface = gr.Interface(
|
| 40 |
+
fn=classify,
|
| 41 |
+
inputs=input_components,
|
| 42 |
+
outputs=gr.Label(num_top_classes=2),
|
| 43 |
+
title="Breast Cancer Classifier",
|
| 44 |
+
description="Enter the required measurements to predict the classification for breast cancer."
|
| 45 |
+
)
|
| 46 |
+
|
| 47 |
+
# Launch the Gradio app
|
| 48 |
+
iface.launch()
|
configs/deployment.yaml
ADDED
|
@@ -0,0 +1,13 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# environment configuration
|
| 2 |
+
settings:
|
| 3 |
+
docker:
|
| 4 |
+
required_integrations:
|
| 5 |
+
- sklearn
|
| 6 |
+
|
| 7 |
+
# configuration of the Model Control Plane
|
| 8 |
+
model_version:
|
| 9 |
+
name: breast_cancer_classifier
|
| 10 |
+
version: production
|
| 11 |
+
license: Apache 2.0
|
| 12 |
+
description: Classification of Breast Cancer Dataset.
|
| 13 |
+
tags: ["classification", "sklearn"]
|
configs/feature_engineering.yaml
ADDED
|
@@ -0,0 +1,12 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# environment configuration
|
| 2 |
+
settings:
|
| 3 |
+
docker:
|
| 4 |
+
required_integrations:
|
| 5 |
+
- sklearn
|
| 6 |
+
|
| 7 |
+
# configuration of the Model Control Plane
|
| 8 |
+
model_version:
|
| 9 |
+
name: breast_cancer_classifier
|
| 10 |
+
license: Apache 2.0
|
| 11 |
+
description: Classification of Breast Cancer Dataset.
|
| 12 |
+
tags: ["classification", "sklearn"]
|
configs/inference.yaml
ADDED
|
@@ -0,0 +1,13 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# environment configuration
|
| 2 |
+
settings:
|
| 3 |
+
docker:
|
| 4 |
+
required_integrations:
|
| 5 |
+
- sklearn
|
| 6 |
+
|
| 7 |
+
# configuration of the Model Control Plane
|
| 8 |
+
model_version:
|
| 9 |
+
name: breast_cancer_classifier
|
| 10 |
+
version: production
|
| 11 |
+
license: Apache 2.0
|
| 12 |
+
description: Classification of Breast Cancer Dataset.
|
| 13 |
+
tags: ["classification", "sklearn"]
|
configs/training.yaml
ADDED
|
@@ -0,0 +1,12 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# environment configuration
|
| 2 |
+
settings:
|
| 3 |
+
docker:
|
| 4 |
+
required_integrations:
|
| 5 |
+
- sklearn
|
| 6 |
+
|
| 7 |
+
# configuration of the Model Control Plane
|
| 8 |
+
model_version:
|
| 9 |
+
name: breast_cancer_classifier
|
| 10 |
+
license: Apache 2.0
|
| 11 |
+
description: Classification of Breast Cancer Dataset.
|
| 12 |
+
tags: ["classification", "sklearn"]
|
flagged/log.csv
ADDED
|
@@ -0,0 +1,2 @@
|
|
|
|
|
|
|
|
|
|
| 1 |
+
mean radius,mean texture,mean perimeter,mean area,mean smoothness,mean compactness,mean concavity,mean concave points,mean symmetry,mean fractal dimension,radius error,texture error,perimeter error,area error,smoothness error,compactness error,concavity error,concave points error,symmetry error,fractal dimension error,worst radius,worst texture,worst perimeter,worst area,worst smoothness,worst compactness,worst concavity,worst concave points,worst symmetry,worst fractal dimension,output,flag,username,timestamp
|
| 2 |
+
2,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,/home/htahir1/workspace/zenml_io/zenml-projects/stack-showcase/flagged/output/tmpjy2eamkw.json,,,2024-01-04 14:08:33.097778
|
flagged/output/tmpjy2eamkw.json
ADDED
|
@@ -0,0 +1 @@
|
|
|
|
|
|
|
| 1 |
+
{}
|
pipelines/__init__.py
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# {% include 'template/license_header' %}
|
| 2 |
+
|
| 3 |
+
from .feature_engineering import feature_engineering
|
| 4 |
+
from .inference import inference
|
| 5 |
+
from .training import breast_cancer_training
|
| 6 |
+
from .deployment import breast_cancer_deployment_pipeline
|
pipelines/__pycache__/__init__.cpython-38.pyc
ADDED
|
Binary file (372 Bytes). View file
|
|
|
pipelines/__pycache__/deployment.cpython-38.pyc
ADDED
|
Binary file (1.3 kB). View file
|
|
|
pipelines/__pycache__/feature_engineering.cpython-38.pyc
ADDED
|
Binary file (1.47 kB). View file
|
|
|
pipelines/__pycache__/inference.cpython-38.pyc
ADDED
|
Binary file (1.43 kB). View file
|
|
|
pipelines/__pycache__/training.cpython-38.pyc
ADDED
|
Binary file (1.55 kB). View file
|
|
|
pipelines/deployment.py
ADDED
|
@@ -0,0 +1,38 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# {% include 'template/license_header' %}
|
| 2 |
+
|
| 3 |
+
from typing import Optional, List
|
| 4 |
+
|
| 5 |
+
from steps import (
|
| 6 |
+
deploy_to_huggingface,
|
| 7 |
+
)
|
| 8 |
+
from zenml import get_pipeline_context, pipeline
|
| 9 |
+
from zenml.logger import get_logger
|
| 10 |
+
from zenml.client import Client
|
| 11 |
+
|
| 12 |
+
logger = get_logger(__name__)
|
| 13 |
+
|
| 14 |
+
|
| 15 |
+
@pipeline
|
| 16 |
+
def breast_cancer_deployment_pipeline(
|
| 17 |
+
repo_name: Optional[str] = "zenml_breast_cancer_classifier",
|
| 18 |
+
):
|
| 19 |
+
"""
|
| 20 |
+
Model deployment pipeline.
|
| 21 |
+
|
| 22 |
+
This pipelines deploys latest model on mlflow registry that matches
|
| 23 |
+
the given stage, to one of the supported deployment targets.
|
| 24 |
+
|
| 25 |
+
Args:
|
| 26 |
+
labels: List of labels for the model.
|
| 27 |
+
title: Title for the model.
|
| 28 |
+
description: Description for the model.
|
| 29 |
+
model_name_or_path: Name or path of the model.
|
| 30 |
+
tokenizer_name_or_path: Name or path of the tokenizer.
|
| 31 |
+
interpretation: Interpretation for the model.
|
| 32 |
+
example: Example for the model.
|
| 33 |
+
repo_name: Name of the repository to deploy to HuggingFace Hub.
|
| 34 |
+
"""
|
| 35 |
+
########## Deploy to HuggingFace ##########
|
| 36 |
+
deploy_to_huggingface(
|
| 37 |
+
repo_name=repo_name,
|
| 38 |
+
)
|
pipelines/feature_engineering.py
ADDED
|
@@ -0,0 +1,54 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# {% include 'template/license_header' %}
|
| 2 |
+
|
| 3 |
+
import random
|
| 4 |
+
from typing import List, Optional
|
| 5 |
+
|
| 6 |
+
from steps import (
|
| 7 |
+
data_loader,
|
| 8 |
+
data_preprocessor,
|
| 9 |
+
data_splitter,
|
| 10 |
+
)
|
| 11 |
+
from zenml import pipeline
|
| 12 |
+
from zenml.logger import get_logger
|
| 13 |
+
|
| 14 |
+
logger = get_logger(__name__)
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
@pipeline
|
| 18 |
+
def feature_engineering(
|
| 19 |
+
test_size: float = 0.2,
|
| 20 |
+
drop_na: Optional[bool] = None,
|
| 21 |
+
normalize: Optional[bool] = None,
|
| 22 |
+
drop_columns: Optional[List[str]] = None,
|
| 23 |
+
target: Optional[str] = "target",
|
| 24 |
+
):
|
| 25 |
+
"""
|
| 26 |
+
Feature engineering pipeline.
|
| 27 |
+
|
| 28 |
+
This is a pipeline that loads the data, processes it and splits
|
| 29 |
+
it into train and test sets.
|
| 30 |
+
|
| 31 |
+
Args:
|
| 32 |
+
test_size: Size of holdout set for training 0.0..1.0
|
| 33 |
+
drop_na: If `True` NA values will be removed from dataset
|
| 34 |
+
normalize: If `True` dataset will be normalized with MinMaxScaler
|
| 35 |
+
drop_columns: List of columns to drop from dataset
|
| 36 |
+
target: Name of target column in dataset
|
| 37 |
+
"""
|
| 38 |
+
### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###
|
| 39 |
+
# Link all the steps together by calling them and passing the output
|
| 40 |
+
# of one step as the input of the next step.
|
| 41 |
+
raw_data = data_loader(random_state=random.randint(0, 100), target=target)
|
| 42 |
+
dataset_trn, dataset_tst = data_splitter(
|
| 43 |
+
dataset=raw_data,
|
| 44 |
+
test_size=test_size,
|
| 45 |
+
)
|
| 46 |
+
dataset_trn, dataset_tst, _ = data_preprocessor(
|
| 47 |
+
dataset_trn=dataset_trn,
|
| 48 |
+
dataset_tst=dataset_tst,
|
| 49 |
+
drop_na=drop_na,
|
| 50 |
+
normalize=normalize,
|
| 51 |
+
drop_columns=drop_columns,
|
| 52 |
+
target=target,
|
| 53 |
+
)
|
| 54 |
+
return dataset_trn, dataset_tst
|
pipelines/inference.py
ADDED
|
@@ -0,0 +1,50 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# {% include 'template/license_header' %}
|
| 2 |
+
|
| 3 |
+
from typing import List, Optional
|
| 4 |
+
|
| 5 |
+
from steps import (
|
| 6 |
+
data_loader,
|
| 7 |
+
inference_preprocessor,
|
| 8 |
+
inference_predict,
|
| 9 |
+
)
|
| 10 |
+
from zenml import pipeline, ExternalArtifact
|
| 11 |
+
from zenml.logger import get_logger
|
| 12 |
+
|
| 13 |
+
logger = get_logger(__name__)
|
| 14 |
+
|
| 15 |
+
|
| 16 |
+
@pipeline
|
| 17 |
+
def inference(
|
| 18 |
+
test_size: float = 0.2,
|
| 19 |
+
drop_na: Optional[bool] = None,
|
| 20 |
+
normalize: Optional[bool] = None,
|
| 21 |
+
drop_columns: Optional[List[str]] = None,
|
| 22 |
+
):
|
| 23 |
+
"""
|
| 24 |
+
Model training pipeline.
|
| 25 |
+
|
| 26 |
+
This is a pipeline that loads the data, processes it and splits
|
| 27 |
+
it into train and test sets, then search for best hyperparameters,
|
| 28 |
+
trains and evaluates a model.
|
| 29 |
+
|
| 30 |
+
Args:
|
| 31 |
+
test_size: Size of holdout set for training 0.0..1.0
|
| 32 |
+
drop_na: If `True` NA values will be removed from dataset
|
| 33 |
+
normalize: If `True` dataset will be normalized with MinMaxScaler
|
| 34 |
+
drop_columns: List of columns to drop from dataset
|
| 35 |
+
"""
|
| 36 |
+
### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###
|
| 37 |
+
# Link all the steps together by calling them and passing the output
|
| 38 |
+
# of one step as the input of the next step.
|
| 39 |
+
random_state = 60
|
| 40 |
+
target = "target"
|
| 41 |
+
df_inference = data_loader(random_state=random_state, is_inference=True)
|
| 42 |
+
df_inference = inference_preprocessor(
|
| 43 |
+
dataset_inf=df_inference,
|
| 44 |
+
preprocess_pipeline=ExternalArtifact(name="preprocess_pipeline"),
|
| 45 |
+
target=target,
|
| 46 |
+
)
|
| 47 |
+
inference_predict(
|
| 48 |
+
dataset_inf=df_inference,
|
| 49 |
+
)
|
| 50 |
+
### END CODE HERE ###
|
pipelines/training.py
ADDED
|
@@ -0,0 +1,61 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# {% include 'template/license_header' %}
|
| 2 |
+
|
| 3 |
+
from typing import Optional
|
| 4 |
+
from uuid import UUID
|
| 5 |
+
|
| 6 |
+
from steps import model_evaluator, model_trainer, model_promoter
|
| 7 |
+
from zenml import ExternalArtifact, pipeline
|
| 8 |
+
from zenml.logger import get_logger
|
| 9 |
+
|
| 10 |
+
from pipelines import (
|
| 11 |
+
feature_engineering,
|
| 12 |
+
)
|
| 13 |
+
|
| 14 |
+
logger = get_logger(__name__)
|
| 15 |
+
|
| 16 |
+
|
| 17 |
+
@pipeline(enable_cache=True)
|
| 18 |
+
def breast_cancer_training(
|
| 19 |
+
train_dataset_id: Optional[UUID] = None,
|
| 20 |
+
test_dataset_id: Optional[UUID] = None,
|
| 21 |
+
min_train_accuracy: float = 0.0,
|
| 22 |
+
min_test_accuracy: float = 0.0,
|
| 23 |
+
):
|
| 24 |
+
"""
|
| 25 |
+
Model training pipeline.
|
| 26 |
+
|
| 27 |
+
This is a pipeline that loads the data, processes it and splits
|
| 28 |
+
it into train and test sets, then search for best hyperparameters,
|
| 29 |
+
trains and evaluates a model.
|
| 30 |
+
|
| 31 |
+
Args:
|
| 32 |
+
test_size: Size of holdout set for training 0.0..1.0
|
| 33 |
+
drop_na: If `True` NA values will be removed from dataset
|
| 34 |
+
normalize: If `True` dataset will be normalized with MinMaxScaler
|
| 35 |
+
drop_columns: List of columns to drop from dataset
|
| 36 |
+
"""
|
| 37 |
+
### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###
|
| 38 |
+
# Link all the steps together by calling them and passing the output
|
| 39 |
+
# of one step as the input of the next step.
|
| 40 |
+
|
| 41 |
+
# Execute Feature Engineering Pipeline
|
| 42 |
+
if train_dataset_id is None or test_dataset_id is None:
|
| 43 |
+
dataset_trn, dataset_tst = feature_engineering()
|
| 44 |
+
else:
|
| 45 |
+
dataset_trn = ExternalArtifact(id=train_dataset_id)
|
| 46 |
+
dataset_tst = ExternalArtifact(id=test_dataset_id)
|
| 47 |
+
|
| 48 |
+
model = model_trainer(
|
| 49 |
+
dataset_trn=dataset_trn,
|
| 50 |
+
)
|
| 51 |
+
|
| 52 |
+
acc = model_evaluator(
|
| 53 |
+
model=model,
|
| 54 |
+
dataset_trn=dataset_trn,
|
| 55 |
+
dataset_tst=dataset_tst,
|
| 56 |
+
min_train_accuracy=min_train_accuracy,
|
| 57 |
+
min_test_accuracy=min_test_accuracy,
|
| 58 |
+
)
|
| 59 |
+
|
| 60 |
+
model_promoter(accuracy=acc)
|
| 61 |
+
### END CODE HERE ###
|
requirements.txt
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
zenml[server]>=0.50.0
|
| 2 |
+
notebook
|
| 3 |
+
scikit-learn<1.3
|
run.ipynb
ADDED
|
@@ -0,0 +1,981 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "code",
|
| 5 |
+
"execution_count": 1,
|
| 6 |
+
"id": "081d5616",
|
| 7 |
+
"metadata": {},
|
| 8 |
+
"outputs": [
|
| 9 |
+
{
|
| 10 |
+
"name": "stdout",
|
| 11 |
+
"output_type": "stream",
|
| 12 |
+
"text": [
|
| 13 |
+
"\u001b[1;35mNumExpr defaulting to 8 threads.\u001b[0m\n",
|
| 14 |
+
"\u001b[?25l\u001b[2;36mFound existing ZenML repository at path \u001b[0m\n",
|
| 15 |
+
"\u001b[2;32m'/home/apenner/PycharmProjects/template-starter/template'\u001b[0m\u001b[2;36m.\u001b[0m\n",
|
| 16 |
+
"\u001b[2;32m⠋\u001b[0m\u001b[2;36m Initializing ZenML repository at \u001b[0m\n",
|
| 17 |
+
"\u001b[2;36m/home/apenner/PycharmProjects/template-starter/template.\u001b[0m\n",
|
| 18 |
+
"\u001b[2K\u001b[1A\u001b[2K\u001b[1A\u001b[2K\u001b[32m⠋\u001b[0m Initializing ZenML repository at \n",
|
| 19 |
+
"/home/apenner/PycharmProjects/template-starter/template.\n",
|
| 20 |
+
"\n",
|
| 21 |
+
"\u001b[1A\u001b[2K\u001b[1A\u001b[2K\u001b[1A\u001b[2K\u001b[1;35mNumExpr defaulting to 8 threads.\u001b[0m\n",
|
| 22 |
+
"\u001b[2K\u001b[2;36mActive repository stack set to: \u001b[0m\u001b[2;32m'default'\u001b[0m.\n",
|
| 23 |
+
"\u001b[2K\u001b[32m⠙\u001b[0m Setting the repository active stack to 'default'...t'...\u001b[0m\n",
|
| 24 |
+
"\u001b[1A\u001b[2K"
|
| 25 |
+
]
|
| 26 |
+
}
|
| 27 |
+
],
|
| 28 |
+
"source": [
|
| 29 |
+
"!zenml init\n",
|
| 30 |
+
"!zenml stack set default"
|
| 31 |
+
]
|
| 32 |
+
},
|
| 33 |
+
{
|
| 34 |
+
"cell_type": "code",
|
| 35 |
+
"execution_count": 2,
|
| 36 |
+
"id": "79f775f2",
|
| 37 |
+
"metadata": {},
|
| 38 |
+
"outputs": [
|
| 39 |
+
{
|
| 40 |
+
"name": "stdout",
|
| 41 |
+
"output_type": "stream",
|
| 42 |
+
"text": [
|
| 43 |
+
"\u001b[1;35mNumExpr defaulting to 8 threads.\u001b[0m\n"
|
| 44 |
+
]
|
| 45 |
+
}
|
| 46 |
+
],
|
| 47 |
+
"source": [
|
| 48 |
+
"# Do the imports at the top\n",
|
| 49 |
+
"\n",
|
| 50 |
+
"import random\n",
|
| 51 |
+
"from zenml import ExternalArtifact, pipeline \n",
|
| 52 |
+
"from zenml.client import Client\n",
|
| 53 |
+
"from zenml.logger import get_logger\n",
|
| 54 |
+
"from uuid import UUID\n",
|
| 55 |
+
"\n",
|
| 56 |
+
"import os\n",
|
| 57 |
+
"from typing import Optional, List\n",
|
| 58 |
+
"\n",
|
| 59 |
+
"from zenml import pipeline\n",
|
| 60 |
+
"\n",
|
| 61 |
+
"from steps import (\n",
|
| 62 |
+
" data_loader,\n",
|
| 63 |
+
" data_preprocessor,\n",
|
| 64 |
+
" data_splitter,\n",
|
| 65 |
+
" model_evaluator,\n",
|
| 66 |
+
" model_trainer,\n",
|
| 67 |
+
" inference_predict,\n",
|
| 68 |
+
" inference_preprocessor\n",
|
| 69 |
+
")\n",
|
| 70 |
+
"\n",
|
| 71 |
+
"logger = get_logger(__name__)\n",
|
| 72 |
+
"\n",
|
| 73 |
+
"client = Client()"
|
| 74 |
+
]
|
| 75 |
+
},
|
| 76 |
+
{
|
| 77 |
+
"cell_type": "code",
|
| 78 |
+
"execution_count": 3,
|
| 79 |
+
"id": "b50a9537",
|
| 80 |
+
"metadata": {},
|
| 81 |
+
"outputs": [],
|
| 82 |
+
"source": [
|
| 83 |
+
"@pipeline\n",
|
| 84 |
+
"def feature_engineering(\n",
|
| 85 |
+
" test_size: float = 0.2,\n",
|
| 86 |
+
" drop_na: Optional[bool] = None,\n",
|
| 87 |
+
" normalize: Optional[bool] = None,\n",
|
| 88 |
+
" drop_columns: Optional[List[str]] = None,\n",
|
| 89 |
+
" target: Optional[str] = \"target\",\n",
|
| 90 |
+
"):\n",
|
| 91 |
+
" \"\"\"\n",
|
| 92 |
+
" Feature engineering pipeline.\n",
|
| 93 |
+
"\n",
|
| 94 |
+
" This is a pipeline that loads the data, processes it and splits\n",
|
| 95 |
+
" it into train and test sets.\n",
|
| 96 |
+
"\n",
|
| 97 |
+
" Args:\n",
|
| 98 |
+
" test_size: Size of holdout set for training 0.0..1.0\n",
|
| 99 |
+
" drop_na: If `True` NA values will be removed from dataset\n",
|
| 100 |
+
" normalize: If `True` dataset will be normalized with MinMaxScaler\n",
|
| 101 |
+
" drop_columns: List of columns to drop from dataset\n",
|
| 102 |
+
" target: Name of target column in dataset\n",
|
| 103 |
+
" \"\"\"\n",
|
| 104 |
+
" ### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###\n",
|
| 105 |
+
" # Link all the steps together by calling them and passing the output\n",
|
| 106 |
+
" # of one step as the input of the next step.\n",
|
| 107 |
+
" raw_data = data_loader(random_state=random.randint(0, 100), target=target)\n",
|
| 108 |
+
" dataset_trn, dataset_tst = data_splitter(\n",
|
| 109 |
+
" dataset=raw_data,\n",
|
| 110 |
+
" test_size=test_size,\n",
|
| 111 |
+
" )\n",
|
| 112 |
+
" dataset_trn, dataset_tst, _ = data_preprocessor(\n",
|
| 113 |
+
" dataset_trn=dataset_trn,\n",
|
| 114 |
+
" dataset_tst=dataset_tst,\n",
|
| 115 |
+
" drop_na=drop_na,\n",
|
| 116 |
+
" normalize=normalize,\n",
|
| 117 |
+
" drop_columns=drop_columns,\n",
|
| 118 |
+
" target=target,\n",
|
| 119 |
+
" )\n",
|
| 120 |
+
" \n",
|
| 121 |
+
" return dataset_trn, dataset_tst"
|
| 122 |
+
]
|
| 123 |
+
},
|
| 124 |
+
{
|
| 125 |
+
"cell_type": "code",
|
| 126 |
+
"execution_count": 4,
|
| 127 |
+
"id": "bc5feef4-7016-420e-9af9-2e87ff666f74",
|
| 128 |
+
"metadata": {},
|
| 129 |
+
"outputs": [],
|
| 130 |
+
"source": [
|
| 131 |
+
"pipeline_args = {}\n",
|
| 132 |
+
"pipeline_args[\"config_path\"] = os.path.join(\"configs\", \"feature_engineering.yaml\")\n",
|
| 133 |
+
"fe_p_configured = feature_engineering.with_options(**pipeline_args)"
|
| 134 |
+
]
|
| 135 |
+
},
|
| 136 |
+
{
|
| 137 |
+
"cell_type": "code",
|
| 138 |
+
"execution_count": 5,
|
| 139 |
+
"id": "75cf3740-b2d8-4c4b-b91b-dc1637000880",
|
| 140 |
+
"metadata": {},
|
| 141 |
+
"outputs": [
|
| 142 |
+
{
|
| 143 |
+
"name": "stdout",
|
| 144 |
+
"output_type": "stream",
|
| 145 |
+
"text": [
|
| 146 |
+
"\u001b[1;35mInitiating a new run for the pipeline: \u001b[0m\u001b[1;36mfeature_engineering\u001b[1;35m.\u001b[0m\n",
|
| 147 |
+
"\u001b[1;35mReusing registered version: \u001b[0m\u001b[1;36m(version: 1)\u001b[1;35m.\u001b[0m\n",
|
| 148 |
+
"\u001b[1;35mNew model version \u001b[0m\u001b[1;36m34\u001b[1;35m was created.\u001b[0m\n",
|
| 149 |
+
"\u001b[1;35mExecuting a new run.\u001b[0m\n",
|
| 150 |
+
"\u001b[1;35mUsing user: \u001b[0m\u001b[1;36malexej@zenml.io\u001b[1;35m\u001b[0m\n",
|
| 151 |
+
"\u001b[1;35mUsing stack: \u001b[0m\u001b[1;36mdefault\u001b[1;35m\u001b[0m\n",
|
| 152 |
+
"\u001b[1;35m artifact_store: \u001b[0m\u001b[1;36mdefault\u001b[1;35m\u001b[0m\n",
|
| 153 |
+
"\u001b[1;35m orchestrator: \u001b[0m\u001b[1;36mdefault\u001b[1;35m\u001b[0m\n",
|
| 154 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mdata_loader\u001b[1;35m has started.\u001b[0m\n",
|
| 155 |
+
"\u001b[1;35mDataset with 541 records loaded!\u001b[0m\n",
|
| 156 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mdata_loader\u001b[1;35m has finished in \u001b[0m\u001b[1;36m6.777s\u001b[1;35m.\u001b[0m\n",
|
| 157 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mdata_splitter\u001b[1;35m has started.\u001b[0m\n",
|
| 158 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mdata_splitter\u001b[1;35m has finished in \u001b[0m\u001b[1;36m11.345s\u001b[1;35m.\u001b[0m\n",
|
| 159 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mdata_preprocessor\u001b[1;35m has started.\u001b[0m\n",
|
| 160 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mdata_preprocessor\u001b[1;35m has finished in \u001b[0m\u001b[1;36m14.866s\u001b[1;35m.\u001b[0m\n",
|
| 161 |
+
"\u001b[1;35mRun \u001b[0m\u001b[1;36mfeature_engineering-2023_12_06-09_08_46_821042\u001b[1;35m has finished in \u001b[0m\u001b[1;36m36.198s\u001b[1;35m.\u001b[0m\n",
|
| 162 |
+
"\u001b[1;35mDashboard URL: https://1cf18d95-zenml.cloudinfra.zenml.io/workspaces/default/pipelines/52874ade-f314-45ab-b9bf-e95fb29290b8/runs/9d9e49b1-d78f-478b-991e-da87b0560512/dag\u001b[0m\n"
|
| 163 |
+
]
|
| 164 |
+
}
|
| 165 |
+
],
|
| 166 |
+
"source": [
|
| 167 |
+
"latest_run = fe_p_configured()"
|
| 168 |
+
]
|
| 169 |
+
},
|
| 170 |
+
{
|
| 171 |
+
"cell_type": "code",
|
| 172 |
+
"execution_count": 6,
|
| 173 |
+
"id": "69ade540",
|
| 174 |
+
"metadata": {},
|
| 175 |
+
"outputs": [],
|
| 176 |
+
"source": [
|
| 177 |
+
"@pipeline\n",
|
| 178 |
+
"def training(\n",
|
| 179 |
+
" train_dataset_id: Optional[UUID] = None,\n",
|
| 180 |
+
" test_dataset_id: Optional[UUID] = None,\n",
|
| 181 |
+
" min_train_accuracy: float = 0.0,\n",
|
| 182 |
+
" min_test_accuracy: float = 0.0,\n",
|
| 183 |
+
"):\n",
|
| 184 |
+
" \"\"\"\n",
|
| 185 |
+
" Model training pipeline.\n",
|
| 186 |
+
"\n",
|
| 187 |
+
" This is a pipeline that loads the data, processes it and splits\n",
|
| 188 |
+
" it into train and test sets, then search for best hyperparameters,\n",
|
| 189 |
+
" trains and evaluates a model.\n",
|
| 190 |
+
"\n",
|
| 191 |
+
" Args:\n",
|
| 192 |
+
" test_size: Size of holdout set for training 0.0..1.0\n",
|
| 193 |
+
" drop_na: If `True` NA values will be removed from dataset\n",
|
| 194 |
+
" normalize: If `True` dataset will be normalized with MinMaxScaler\n",
|
| 195 |
+
" drop_columns: List of columns to drop from dataset\n",
|
| 196 |
+
" \"\"\"\n",
|
| 197 |
+
" ### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###\n",
|
| 198 |
+
" # Link all the steps together by calling them and passing the output\n",
|
| 199 |
+
" # of one step as the input of the next step.\n",
|
| 200 |
+
" \n",
|
| 201 |
+
" # Execute Feature Engineering Pipeline\n",
|
| 202 |
+
" if train_dataset_id is None or test_dataset_id is None:\n",
|
| 203 |
+
" dataset_trn, dataset_tst = feature_engineering()\n",
|
| 204 |
+
" else:\n",
|
| 205 |
+
" dataset_trn = ExternalArtifact(id=train_dataset_id)\n",
|
| 206 |
+
" dataset_tst = ExternalArtifact(id=test_dataset_id)\n",
|
| 207 |
+
" \n",
|
| 208 |
+
" model = model_trainer(\n",
|
| 209 |
+
" dataset_trn=dataset_trn,\n",
|
| 210 |
+
" )\n",
|
| 211 |
+
"\n",
|
| 212 |
+
" model_evaluator(\n",
|
| 213 |
+
" model=model,\n",
|
| 214 |
+
" dataset_trn=dataset_trn,\n",
|
| 215 |
+
" dataset_tst=dataset_tst,\n",
|
| 216 |
+
" min_train_accuracy=min_train_accuracy,\n",
|
| 217 |
+
" min_test_accuracy=min_test_accuracy,\n",
|
| 218 |
+
" )\n"
|
| 219 |
+
]
|
| 220 |
+
},
|
| 221 |
+
{
|
| 222 |
+
"cell_type": "code",
|
| 223 |
+
"execution_count": 7,
|
| 224 |
+
"id": "5b1f78df",
|
| 225 |
+
"metadata": {},
|
| 226 |
+
"outputs": [],
|
| 227 |
+
"source": [
|
| 228 |
+
"pipeline_args = {}\n",
|
| 229 |
+
"pipeline_args[\"config_path\"] = os.path.join(\"configs\", \"training.yaml\")\n",
|
| 230 |
+
"fe_t_configured = training.with_options(**pipeline_args)"
|
| 231 |
+
]
|
| 232 |
+
},
|
| 233 |
+
{
|
| 234 |
+
"cell_type": "code",
|
| 235 |
+
"execution_count": 8,
|
| 236 |
+
"id": "acf306a5",
|
| 237 |
+
"metadata": {},
|
| 238 |
+
"outputs": [
|
| 239 |
+
{
|
| 240 |
+
"name": "stdout",
|
| 241 |
+
"output_type": "stream",
|
| 242 |
+
"text": [
|
| 243 |
+
"\u001b[1;35mInitiating a new run for the pipeline: \u001b[0m\u001b[1;36mtraining\u001b[1;35m.\u001b[0m\n",
|
| 244 |
+
"\u001b[1;35mRegistered new version: \u001b[0m\u001b[1;36m(version 2)\u001b[1;35m.\u001b[0m\n",
|
| 245 |
+
"\u001b[1;35mNew model version \u001b[0m\u001b[1;36m35\u001b[1;35m was created.\u001b[0m\n",
|
| 246 |
+
"\u001b[1;35mExecuting a new run.\u001b[0m\n",
|
| 247 |
+
"\u001b[1;35mUsing user: \u001b[0m\u001b[1;36malexej@zenml.io\u001b[1;35m\u001b[0m\n",
|
| 248 |
+
"\u001b[1;35mUsing stack: \u001b[0m\u001b[1;36mdefault\u001b[1;35m\u001b[0m\n",
|
| 249 |
+
"\u001b[1;35m artifact_store: \u001b[0m\u001b[1;36mdefault\u001b[1;35m\u001b[0m\n",
|
| 250 |
+
"\u001b[1;35m orchestrator: \u001b[0m\u001b[1;36mdefault\u001b[1;35m\u001b[0m\n",
|
| 251 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mdata_loader\u001b[1;35m has started.\u001b[0m\n",
|
| 252 |
+
"\u001b[1;35mDataset with 541 records loaded!\u001b[0m\n",
|
| 253 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mdata_loader\u001b[1;35m has finished in \u001b[0m\u001b[1;36m7.368s\u001b[1;35m.\u001b[0m\n",
|
| 254 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mdata_splitter\u001b[1;35m has started.\u001b[0m\n",
|
| 255 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mdata_splitter\u001b[1;35m has finished in \u001b[0m\u001b[1;36m11.009s\u001b[1;35m.\u001b[0m\n",
|
| 256 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mdata_preprocessor\u001b[1;35m has started.\u001b[0m\n",
|
| 257 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mdata_preprocessor\u001b[1;35m has finished in \u001b[0m\u001b[1;36m14.134s\u001b[1;35m.\u001b[0m\n",
|
| 258 |
+
"\u001b[1;35mCaching \u001b[0m\u001b[1;36mdisabled\u001b[1;35m explicitly for \u001b[0m\u001b[1;36mmodel_trainer\u001b[1;35m.\u001b[0m\n",
|
| 259 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mmodel_trainer\u001b[1;35m has started.\u001b[0m\n",
|
| 260 |
+
"\u001b[1;35mTraining model DecisionTreeClassifier()...\u001b[0m\n",
|
| 261 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mmodel_trainer\u001b[1;35m has finished in \u001b[0m\u001b[1;36m7.035s\u001b[1;35m.\u001b[0m\n",
|
| 262 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mmodel_evaluator\u001b[1;35m has started.\u001b[0m\n",
|
| 263 |
+
"\u001b[1;35mTrain accuracy=100.00%\u001b[0m\n",
|
| 264 |
+
"\u001b[1;35mTest accuracy=92.66%\u001b[0m\n",
|
| 265 |
+
"\u001b[1;35mImplicitly linking artifact \u001b[0m\u001b[1;36moutput\u001b[1;35m to model \u001b[0m\u001b[1;36mbreast_cancer_classifier\u001b[1;35m version \u001b[0m\u001b[1;36m35\u001b[1;35m.\u001b[0m\n",
|
| 266 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mmodel_evaluator\u001b[1;35m has finished in \u001b[0m\u001b[1;36m6.050s\u001b[1;35m.\u001b[0m\n",
|
| 267 |
+
"\u001b[1;35mRun \u001b[0m\u001b[1;36mtraining-2023_12_06-09_09_41_413455\u001b[1;35m has finished in \u001b[0m\u001b[1;36m51.278s\u001b[1;35m.\u001b[0m\n",
|
| 268 |
+
"\u001b[1;35mDashboard URL: https://1cf18d95-zenml.cloudinfra.zenml.io/workspaces/default/pipelines/787c6360-4499-4e2e-8d50-edaaa3956a6f/runs/2a335b9c-bb8e-425c-80e2-0a6cc0ffe56a/dag\u001b[0m\n"
|
| 269 |
+
]
|
| 270 |
+
}
|
| 271 |
+
],
|
| 272 |
+
"source": [
|
| 273 |
+
"fe_t_configured()"
|
| 274 |
+
]
|
| 275 |
+
},
|
| 276 |
+
{
|
| 277 |
+
"cell_type": "code",
|
| 278 |
+
"execution_count": 9,
|
| 279 |
+
"id": "ad6aa280",
|
| 280 |
+
"metadata": {},
|
| 281 |
+
"outputs": [],
|
| 282 |
+
"source": [
|
| 283 |
+
"from typing import Optional\n",
|
| 284 |
+
"\n",
|
| 285 |
+
"import pandas as pd\n",
|
| 286 |
+
"from typing_extensions import Annotated\n",
|
| 287 |
+
"\n",
|
| 288 |
+
"from zenml import get_step_context, step\n",
|
| 289 |
+
"from zenml.logger import get_logger\n",
|
| 290 |
+
"\n",
|
| 291 |
+
"logger = get_logger(__name__)\n",
|
| 292 |
+
"\n",
|
| 293 |
+
"\n",
|
| 294 |
+
"@step\n",
|
| 295 |
+
"def inference_predict(\n",
|
| 296 |
+
" dataset_inf: pd.DataFrame,\n",
|
| 297 |
+
") -> Annotated[pd.Series, \"predictions\"]:\n",
|
| 298 |
+
" \"\"\"Predictions step.\n",
|
| 299 |
+
"\n",
|
| 300 |
+
" This is an example of a predictions step that takes the data in and returns\n",
|
| 301 |
+
" predicted values.\n",
|
| 302 |
+
"\n",
|
| 303 |
+
" This step is parameterized, which allows you to configure the step\n",
|
| 304 |
+
" independently of the step code, before running it in a pipeline.\n",
|
| 305 |
+
" In this example, the step can be configured to use different input data.\n",
|
| 306 |
+
" See the documentation for more information:\n",
|
| 307 |
+
"\n",
|
| 308 |
+
" https://docs.zenml.io/user-guide/advanced-guide/configure-steps-pipelines\n",
|
| 309 |
+
"\n",
|
| 310 |
+
" Args:\n",
|
| 311 |
+
" dataset_inf: The inference dataset.\n",
|
| 312 |
+
"\n",
|
| 313 |
+
" Returns:\n",
|
| 314 |
+
" The predictions as pandas series\n",
|
| 315 |
+
" \"\"\"\n",
|
| 316 |
+
" ### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###\n",
|
| 317 |
+
" model_version = get_step_context().model_version\n",
|
| 318 |
+
"\n",
|
| 319 |
+
" print(model_version)\n",
|
| 320 |
+
"\n",
|
| 321 |
+
" # run prediction from memory\n",
|
| 322 |
+
" predictor = model_version.load_artifact(\"model\")\n",
|
| 323 |
+
" predictions = predictor.predict(dataset_inf)\n",
|
| 324 |
+
"\n",
|
| 325 |
+
" print(predictions)\n",
|
| 326 |
+
" predictions = pd.Series(predictions, name=\"predicted\")\n",
|
| 327 |
+
" ### YOUR CODE ENDS HERE ###\n",
|
| 328 |
+
"\n",
|
| 329 |
+
" return predictions\n"
|
| 330 |
+
]
|
| 331 |
+
},
|
| 332 |
+
{
|
| 333 |
+
"cell_type": "code",
|
| 334 |
+
"execution_count": 10,
|
| 335 |
+
"id": "517ad39d",
|
| 336 |
+
"metadata": {},
|
| 337 |
+
"outputs": [],
|
| 338 |
+
"source": [
|
| 339 |
+
"@pipeline\n",
|
| 340 |
+
"def batch_inference():\n",
|
| 341 |
+
" \"\"\"\n",
|
| 342 |
+
" Model batch inference pipeline.\n",
|
| 343 |
+
"\n",
|
| 344 |
+
" This is a pipeline that loads the inference data, processes\n",
|
| 345 |
+
" it, analyze for data drift and run inference.\n",
|
| 346 |
+
" \"\"\"\n",
|
| 347 |
+
" ### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###\n",
|
| 348 |
+
" # Link all the steps together by calling them and passing the output\n",
|
| 349 |
+
" # of one step as the input of the next step.\n",
|
| 350 |
+
" ########## ETL stage ##########\n",
|
| 351 |
+
" random_state = client.get_artifact(\"dataset\").run_metadata[\"random_state\"].value\n",
|
| 352 |
+
" target = client.get_artifact(\"dataset_trn\").run_metadata['target'].value\n",
|
| 353 |
+
" df_inference = data_loader(\n",
|
| 354 |
+
" random_state=random_state, is_inference=True\n",
|
| 355 |
+
" )\n",
|
| 356 |
+
" df_inference = inference_preprocessor(\n",
|
| 357 |
+
" dataset_inf=df_inference,\n",
|
| 358 |
+
" preprocess_pipeline=ExternalArtifact(name=\"preprocess_pipeline\"),\n",
|
| 359 |
+
" target=target,\n",
|
| 360 |
+
" )\n",
|
| 361 |
+
" inference_predict(\n",
|
| 362 |
+
" dataset_inf=df_inference,\n",
|
| 363 |
+
" )\n"
|
| 364 |
+
]
|
| 365 |
+
},
|
| 366 |
+
{
|
| 367 |
+
"cell_type": "code",
|
| 368 |
+
"execution_count": 11,
|
| 369 |
+
"id": "f0d9ebb6",
|
| 370 |
+
"metadata": {},
|
| 371 |
+
"outputs": [
|
| 372 |
+
{
|
| 373 |
+
"name": "stdout",
|
| 374 |
+
"output_type": "stream",
|
| 375 |
+
"text": [
|
| 376 |
+
"\u001b[1;35m\u001b[0m\u001b[1;36mversion\u001b[1;35m \u001b[0m\u001b[1;36mproduction\u001b[1;35m matches one of the possible \u001b[0m\u001b[1;36mModelStages\u001b[1;35m and will be fetched using stage.\u001b[0m\n"
|
| 377 |
+
]
|
| 378 |
+
}
|
| 379 |
+
],
|
| 380 |
+
"source": [
|
| 381 |
+
"pipeline_args = {}\n",
|
| 382 |
+
"pipeline_args[\"config_path\"] = os.path.join(\"configs\", \"inference.yaml\")\n",
|
| 383 |
+
"fe_b_configured = batch_inference.with_options(**pipeline_args)"
|
| 384 |
+
]
|
| 385 |
+
},
|
| 386 |
+
{
|
| 387 |
+
"cell_type": "code",
|
| 388 |
+
"execution_count": 13,
|
| 389 |
+
"id": "9901c6d0",
|
| 390 |
+
"metadata": {},
|
| 391 |
+
"outputs": [
|
| 392 |
+
{
|
| 393 |
+
"name": "stdout",
|
| 394 |
+
"output_type": "stream",
|
| 395 |
+
"text": [
|
| 396 |
+
"\u001b[33mUsing an external artifact as step input currently invalidates caching for the step and all downstream steps. Future releases will introduce hashing of artifacts which will improve this behavior.\u001b[0m\n",
|
| 397 |
+
"\u001b[1;35mInitiating a new run for the pipeline: \u001b[0m\u001b[1;36mbatch_inference\u001b[1;35m.\u001b[0m\n",
|
| 398 |
+
"\u001b[1;35mReusing registered version: \u001b[0m\u001b[1;36m(version: 1)\u001b[1;35m.\u001b[0m\n",
|
| 399 |
+
"\u001b[1;35mExecuting a new run.\u001b[0m\n",
|
| 400 |
+
"\u001b[1;35mUsing user: \u001b[0m\u001b[1;36malexej@zenml.io\u001b[1;35m\u001b[0m\n",
|
| 401 |
+
"\u001b[1;35mUsing stack: \u001b[0m\u001b[1;36mdefault\u001b[1;35m\u001b[0m\n",
|
| 402 |
+
"\u001b[1;35m artifact_store: \u001b[0m\u001b[1;36mdefault\u001b[1;35m\u001b[0m\n",
|
| 403 |
+
"\u001b[1;35m orchestrator: \u001b[0m\u001b[1;36mdefault\u001b[1;35m\u001b[0m\n",
|
| 404 |
+
"\u001b[1;35mUsing cached version of \u001b[0m\u001b[1;36mdata_loader\u001b[1;35m.\u001b[0m\n",
|
| 405 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36mdata_loader\u001b[1;35m has started.\u001b[0m\n",
|
| 406 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36minference_preprocessor\u001b[1;35m has started.\u001b[0m\n",
|
| 407 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36minference_preprocessor\u001b[1;35m has finished in \u001b[0m\u001b[1;36m8.661s\u001b[1;35m.\u001b[0m\n",
|
| 408 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36minference_predict\u001b[1;35m has started.\u001b[0m\n",
|
| 409 |
+
"name='breast_cancer_classifier' license='Apache 2.0' description='Classification of Breast Cancer Dataset.' audience=None use_cases=None limitations=None trade_offs=None ethics=None tags=['classification', 'sklearn'] version='production' save_models_to_registry=True suppress_class_validation_warnings=True was_created_in_this_run=False\n",
|
| 410 |
+
"\u001b[33mYou specified both an ID as well as a version of the artifacts. Ignoring the version and fetching the artifacts by ID.\u001b[0m\n",
|
| 411 |
+
"\u001b[33mYour artifact was materialized under Python version 'unknown' but you are currently using '3.9.13'. This might cause unexpected behavior since pickle is not reproducible across Python versions. Attempting to load anyway...\u001b[0m\n",
|
| 412 |
+
"\u001b[33mCould not import Azure service connector: No module named 'azure.identity'.\u001b[0m\n",
|
| 413 |
+
"[1 0 0 1 1 0 0 0 0 1 1 0 1 0 1 0 1 1 1 0 0 1 0 1 1 1 1 1]\n",
|
| 414 |
+
"\u001b[1;35mStep \u001b[0m\u001b[1;36minference_predict\u001b[1;35m has finished in \u001b[0m\u001b[1;36m18.218s\u001b[1;35m.\u001b[0m\n",
|
| 415 |
+
"\u001b[1;35mRun \u001b[0m\u001b[1;36mbatch_inference-2023_12_06-09_11_29_924914\u001b[1;35m has finished in \u001b[0m\u001b[1;36m32.726s\u001b[1;35m.\u001b[0m\n",
|
| 416 |
+
"\u001b[1;35mDashboard URL: https://1cf18d95-zenml.cloudinfra.zenml.io/workspaces/default/pipelines/2979acb2-c862-480a-8f50-a2be4c76a8a2/runs/7886e370-b05a-4205-931e-e4994fabd897/dag\u001b[0m\n"
|
| 417 |
+
]
|
| 418 |
+
}
|
| 419 |
+
],
|
| 420 |
+
"source": [
|
| 421 |
+
"fe_b_configured()"
|
| 422 |
+
]
|
| 423 |
+
},
|
| 424 |
+
{
|
| 425 |
+
"cell_type": "code",
|
| 426 |
+
"execution_count": null,
|
| 427 |
+
"id": "98d39df8",
|
| 428 |
+
"metadata": {},
|
| 429 |
+
"outputs": [],
|
| 430 |
+
"source": []
|
| 431 |
+
},
|
| 432 |
+
{
|
| 433 |
+
"cell_type": "markdown",
|
| 434 |
+
"id": "51690802-31a7-4e6d-9f88-e6457c6c4a96",
|
| 435 |
+
"metadata": {},
|
| 436 |
+
"source": [
|
| 437 |
+
"# Huggingface Model to Sagemaker Endpoint: Automating MLOps with ZenML\n",
|
| 438 |
+
"Deploying Huggingface models to AWS Sagemaker endpoints typically only requires a few lines of code. However, there's a growing demand to not just deploy, but to seamlessly automate the entire flow from training to production with comprehensive lineage tracking. ZenML adeptly fills this niche, providing an end-to-end MLOps solution for Huggingface users wishing to deploy to Sagemaker. Below, we’ll walk through the architecture that ZenML employs to bring a Huggingface model into production with AWS Sagemaker. Of course all of this can be adapted to not just Sagemaker, but any other model deployment service like GCP Vertex or Azure ML Platform.\n",
|
| 439 |
+
"\n",
|
| 440 |
+
"This blog post showcases one way of using ZenML pipelines to achieve this:\n",
|
| 441 |
+
"\n",
|
| 442 |
+
"- Create and version a dataset in a feature_engineering_pipeline.\n",
|
| 443 |
+
"- Train/Finetune a BERT-based Sentiment Analysis NLP model and push to Huggingface Hub in a training_pipeline.\n",
|
| 444 |
+
"- Promote this model to Production by comparing to previous models in a promotion_pipeline.\n",
|
| 445 |
+
"- Deploy the model at the Production Stage to a AWS Sagemaker endpoint with a deployment_pipeline.\n",
|
| 446 |
+
"\n",
|
| 447 |
+
"<img src=\"assets/pipelines_overview.png\" alt=\"Pipelines Overview\">"
|
| 448 |
+
]
|
| 449 |
+
},
|
| 450 |
+
{
|
| 451 |
+
"cell_type": "code",
|
| 452 |
+
"execution_count": null,
|
| 453 |
+
"id": "500e3c24-b105-4a69-b2fc-e0ce1f1c1d46",
|
| 454 |
+
"metadata": {},
|
| 455 |
+
"outputs": [],
|
| 456 |
+
"source": [
|
| 457 |
+
"# Do the imports at the top\n",
|
| 458 |
+
"\n",
|
| 459 |
+
"import numpy as np\n",
|
| 460 |
+
"from datasets import DatasetDict, load_dataset\n",
|
| 461 |
+
"from typing_extensions import Annotated\n",
|
| 462 |
+
"from zenml import step\n",
|
| 463 |
+
"from zenml.logger import get_logger\n",
|
| 464 |
+
"\n",
|
| 465 |
+
"import os\n",
|
| 466 |
+
"from typing import Optional\n",
|
| 467 |
+
"from datetime import datetime as dt\n",
|
| 468 |
+
"\n",
|
| 469 |
+
"from zenml import pipeline\n",
|
| 470 |
+
"from zenml.model import ModelConfig\n",
|
| 471 |
+
"\n",
|
| 472 |
+
"from steps import (\n",
|
| 473 |
+
" data_loader,\n",
|
| 474 |
+
" notify_on_failure,\n",
|
| 475 |
+
" tokenization_step,\n",
|
| 476 |
+
" tokenizer_loader,\n",
|
| 477 |
+
" generate_reference_and_comparison_datasets,\n",
|
| 478 |
+
")\n",
|
| 479 |
+
"from zenml.integrations.evidently.metrics import EvidentlyMetricConfig\n",
|
| 480 |
+
"from zenml.integrations.evidently.steps import (\n",
|
| 481 |
+
" EvidentlyColumnMapping,\n",
|
| 482 |
+
" evidently_report_step,\n",
|
| 483 |
+
")\n",
|
| 484 |
+
"\n",
|
| 485 |
+
"from pipelines import (\n",
|
| 486 |
+
" sentinment_analysis_deploy_pipeline,\n",
|
| 487 |
+
" sentinment_analysis_promote_pipeline,\n",
|
| 488 |
+
" sentinment_analysis_training_pipeline,\n",
|
| 489 |
+
")\n",
|
| 490 |
+
"\n",
|
| 491 |
+
"logger = get_logger(__name__)"
|
| 492 |
+
]
|
| 493 |
+
},
|
| 494 |
+
{
|
| 495 |
+
"cell_type": "markdown",
|
| 496 |
+
"id": "fc77b660-e206-46b1-a924-407e797a8f47",
|
| 497 |
+
"metadata": {},
|
| 498 |
+
"source": [
|
| 499 |
+
"# 🍳Breaking it down\n",
|
| 500 |
+
"\n",
|
| 501 |
+
"\n",
|
| 502 |
+
"\n"
|
| 503 |
+
]
|
| 504 |
+
},
|
| 505 |
+
{
|
| 506 |
+
"cell_type": "markdown",
|
| 507 |
+
"id": "31edaf46-6981-42be-99b7-9bdd91c160d5",
|
| 508 |
+
"metadata": {},
|
| 509 |
+
"source": [
|
| 510 |
+
"## 👶 Step 1: Start with feature engineering\n",
|
| 511 |
+
"\n",
|
| 512 |
+
"Automated feature engineering forms the foundation of this MLOps workflow. Thats why the first pipeline is the feature engineering pipeline. This pipeline loads some data from Huggingface and uses a base tokenizer to create a tokenized dataset. The data loader step is a simple Python function that returns a Huggingface dataloader object:"
|
| 513 |
+
]
|
| 514 |
+
},
|
| 515 |
+
{
|
| 516 |
+
"cell_type": "code",
|
| 517 |
+
"execution_count": null,
|
| 518 |
+
"id": "35de0e4c-b6f8-4b68-927a-f40e4130dc93",
|
| 519 |
+
"metadata": {},
|
| 520 |
+
"outputs": [],
|
| 521 |
+
"source": [
|
| 522 |
+
"@step\n",
|
| 523 |
+
"def data_loader() -> Annotated[DatasetDict, \"dataset\"]:\n",
|
| 524 |
+
" logger.info(f\"Loading dataset airline_reviews... \")\n",
|
| 525 |
+
" hf_dataset = load_dataset(\"Shayanvsf/US_Airline_Sentiment\")\n",
|
| 526 |
+
" hf_dataset = hf_dataset.rename_column(\"airline_sentiment\", \"label\")\n",
|
| 527 |
+
" hf_dataset = hf_dataset.remove_columns(\n",
|
| 528 |
+
" [\"airline_sentiment_confidence\", \"negativereason_confidence\"]\n",
|
| 529 |
+
" )\n",
|
| 530 |
+
" return hf_dataset"
|
| 531 |
+
]
|
| 532 |
+
},
|
| 533 |
+
{
|
| 534 |
+
"cell_type": "markdown",
|
| 535 |
+
"id": "49e4462c-1e64-48d3-bae7-76696a958646",
|
| 536 |
+
"metadata": {},
|
| 537 |
+
"source": [
|
| 538 |
+
"Notice that you can give each dataset a name with Python’s Annotated object. The DatasetDict is a native Huggingface dataset which ZenML knows how to persist through steps. This flow ensures reproducibility and version control for every dataset iteration.\n",
|
| 539 |
+
"\n",
|
| 540 |
+
"Also notice this is a simple Python function, that can be called with the `entrypoint` wrapper:"
|
| 541 |
+
]
|
| 542 |
+
},
|
| 543 |
+
{
|
| 544 |
+
"cell_type": "code",
|
| 545 |
+
"execution_count": null,
|
| 546 |
+
"id": "18144a6b-c266-453d-82c8-b5d6aa1be0aa",
|
| 547 |
+
"metadata": {},
|
| 548 |
+
"outputs": [],
|
| 549 |
+
"source": [
|
| 550 |
+
"hf_dataset = data_loader.entrypoint()\n",
|
| 551 |
+
"print(hf_dataset)"
|
| 552 |
+
]
|
| 553 |
+
},
|
| 554 |
+
{
|
| 555 |
+
"cell_type": "markdown",
|
| 556 |
+
"id": "31330d3c-044f-4912-8d36-74146f48cecf",
|
| 557 |
+
"metadata": {},
|
| 558 |
+
"source": [
|
| 559 |
+
"Now we put this a full feature engineering pipeline. Each run of the feature engineering pipeline produces a new dataset to use for the training pipeline. ZenML versions this data as it flows through the pipeline.\n",
|
| 560 |
+
"\n",
|
| 561 |
+
"<img src=\"assets/pipelines_feature_eng.png\" alt=\"Pipelines Feature Engineering\">"
|
| 562 |
+
]
|
| 563 |
+
},
|
| 564 |
+
{
|
| 565 |
+
"cell_type": "markdown",
|
| 566 |
+
"id": "9511bd84-1e97-42db-9b75-06285cc6904c",
|
| 567 |
+
"metadata": {},
|
| 568 |
+
"source": [
|
| 569 |
+
"### Set your stack"
|
| 570 |
+
]
|
| 571 |
+
},
|
| 572 |
+
{
|
| 573 |
+
"cell_type": "code",
|
| 574 |
+
"execution_count": null,
|
| 575 |
+
"id": "76f3a7e7-0d85-43b3-9e9f-4c7f20ea65e6",
|
| 576 |
+
"metadata": {},
|
| 577 |
+
"outputs": [],
|
| 578 |
+
"source": [
|
| 579 |
+
"!zenml stack describe hf-sagemaker-local"
|
| 580 |
+
]
|
| 581 |
+
},
|
| 582 |
+
{
|
| 583 |
+
"cell_type": "code",
|
| 584 |
+
"execution_count": null,
|
| 585 |
+
"id": "04b0bf69-70c6-4408-b18c-95df9e030c0c",
|
| 586 |
+
"metadata": {},
|
| 587 |
+
"outputs": [],
|
| 588 |
+
"source": [
|
| 589 |
+
"!zenml stack set hf-sagemaker-local"
|
| 590 |
+
]
|
| 591 |
+
},
|
| 592 |
+
{
|
| 593 |
+
"cell_type": "code",
|
| 594 |
+
"execution_count": null,
|
| 595 |
+
"id": "de5398a4-a9ec-42d6-bbd6-390244c52d13",
|
| 596 |
+
"metadata": {},
|
| 597 |
+
"outputs": [],
|
| 598 |
+
"source": [
|
| 599 |
+
"!zenml stack get"
|
| 600 |
+
]
|
| 601 |
+
},
|
| 602 |
+
{
|
| 603 |
+
"cell_type": "markdown",
|
| 604 |
+
"id": "152f718d-70c2-4a29-a73e-37db85675cb8",
|
| 605 |
+
"metadata": {},
|
| 606 |
+
"source": [
|
| 607 |
+
"### Run the pipeline"
|
| 608 |
+
]
|
| 609 |
+
},
|
| 610 |
+
{
|
| 611 |
+
"cell_type": "code",
|
| 612 |
+
"execution_count": null,
|
| 613 |
+
"id": "7ca6c41e-e4b3-46d2-8264-9a453ac9aa3c",
|
| 614 |
+
"metadata": {
|
| 615 |
+
"scrolled": true
|
| 616 |
+
},
|
| 617 |
+
"outputs": [],
|
| 618 |
+
"source": [
|
| 619 |
+
"@pipeline(on_failure=notify_on_failure)\n",
|
| 620 |
+
"def sentinment_analysis_feature_engineering_pipeline(\n",
|
| 621 |
+
" lower_case: Optional[bool] = True,\n",
|
| 622 |
+
" padding: Optional[str] = \"max_length\",\n",
|
| 623 |
+
" max_seq_length: Optional[int] = 128,\n",
|
| 624 |
+
" text_column: Optional[str] = \"text\",\n",
|
| 625 |
+
" label_column: Optional[str] = \"label\",\n",
|
| 626 |
+
"):\n",
|
| 627 |
+
" # Link all the steps together by calling them and passing the output\n",
|
| 628 |
+
" # of one step as the input of the next step.\n",
|
| 629 |
+
"\n",
|
| 630 |
+
" ########## Load Dataset stage ##########\n",
|
| 631 |
+
" dataset = data_loader()\n",
|
| 632 |
+
"\n",
|
| 633 |
+
" ########## Data Quality stage ##########\n",
|
| 634 |
+
" reference_dataset, comparison_dataset = generate_reference_and_comparison_datasets(\n",
|
| 635 |
+
" dataset\n",
|
| 636 |
+
" )\n",
|
| 637 |
+
" text_data_report = evidently_report_step.with_options(\n",
|
| 638 |
+
" parameters=dict(\n",
|
| 639 |
+
" column_mapping=EvidentlyColumnMapping(\n",
|
| 640 |
+
" target=\"label\",\n",
|
| 641 |
+
" text_features=[\"text\"],\n",
|
| 642 |
+
" ),\n",
|
| 643 |
+
" metrics=[\n",
|
| 644 |
+
" EvidentlyMetricConfig.metric(\"DataQualityPreset\"),\n",
|
| 645 |
+
" EvidentlyMetricConfig.metric(\n",
|
| 646 |
+
" \"TextOverviewPreset\", column_name=\"text\"\n",
|
| 647 |
+
" ),\n",
|
| 648 |
+
" ],\n",
|
| 649 |
+
" # We need to download the NLTK data for the TextOverviewPreset\n",
|
| 650 |
+
" download_nltk_data=True,\n",
|
| 651 |
+
" ),\n",
|
| 652 |
+
" )\n",
|
| 653 |
+
" text_data_report(reference_dataset, comparison_dataset)\n",
|
| 654 |
+
"\n",
|
| 655 |
+
" ########## Tokenization stage ##########\n",
|
| 656 |
+
" tokenizer = tokenizer_loader(lower_case=lower_case)\n",
|
| 657 |
+
" tokenized_data = tokenization_step(\n",
|
| 658 |
+
" dataset=dataset,\n",
|
| 659 |
+
" tokenizer=tokenizer,\n",
|
| 660 |
+
" padding=padding,\n",
|
| 661 |
+
" max_seq_length=max_seq_length,\n",
|
| 662 |
+
" text_column=text_column,\n",
|
| 663 |
+
" label_column=label_column,\n",
|
| 664 |
+
" )\n",
|
| 665 |
+
" return tokenizer, tokenized_data"
|
| 666 |
+
]
|
| 667 |
+
},
|
| 668 |
+
{
|
| 669 |
+
"cell_type": "code",
|
| 670 |
+
"execution_count": null,
|
| 671 |
+
"id": "3c8a5be7-ebaa-41c4-ac23-4afc6e7e06aa",
|
| 672 |
+
"metadata": {},
|
| 673 |
+
"outputs": [],
|
| 674 |
+
"source": [
|
| 675 |
+
"# Run a pipeline with the required parameters. \n",
|
| 676 |
+
"no_cache: bool = True\n",
|
| 677 |
+
"zenml_model_name: str = \"distil_bert_sentiment_analysis\"\n",
|
| 678 |
+
"max_seq_length = 512\n",
|
| 679 |
+
"\n",
|
| 680 |
+
"# This executes all steps in the pipeline in the correct order using the orchestrator\n",
|
| 681 |
+
"# stack component that is configured in your active ZenML stack.\n",
|
| 682 |
+
"model_config = ModelConfig(\n",
|
| 683 |
+
" name=zenml_model_name,\n",
|
| 684 |
+
" license=\"Apache 2.0\",\n",
|
| 685 |
+
" description=\"Show case Model Control Plane.\",\n",
|
| 686 |
+
" create_new_model_version=True,\n",
|
| 687 |
+
" delete_new_version_on_failure=True,\n",
|
| 688 |
+
" tags=[\"sentiment_analysis\", \"huggingface\"],\n",
|
| 689 |
+
")\n",
|
| 690 |
+
"\n",
|
| 691 |
+
"pipeline_args = {}\n",
|
| 692 |
+
"\n",
|
| 693 |
+
"if no_cache:\n",
|
| 694 |
+
" pipeline_args[\"enable_cache\"] = False\n",
|
| 695 |
+
"\n",
|
| 696 |
+
"# Execute Feature Engineering Pipeline\n",
|
| 697 |
+
"pipeline_args[\"model_config\"] = model_config\n",
|
| 698 |
+
"pipeline_args[\"config_path\"] = os.path.join(\"configs\", \"feature_engineering_config.yaml\")\n",
|
| 699 |
+
"run_args_feature = {\n",
|
| 700 |
+
" \"max_seq_length\": max_seq_length,\n",
|
| 701 |
+
"}\n",
|
| 702 |
+
"pipeline_args[\n",
|
| 703 |
+
" \"run_name\"\n",
|
| 704 |
+
"] = f\"sentinment_analysis_feature_engineering_pipeline_run_{dt.now().strftime('%Y_%m_%d_%H_%M_%S')}\"\n",
|
| 705 |
+
"p = sentinment_analysis_feature_engineering_pipeline.with_options(**pipeline_args)\n",
|
| 706 |
+
"p(**run_args_feature)"
|
| 707 |
+
]
|
| 708 |
+
},
|
| 709 |
+
{
|
| 710 |
+
"cell_type": "code",
|
| 711 |
+
"execution_count": null,
|
| 712 |
+
"id": "0e7c1ea2-64fe-478a-9963-17c7b7f62110",
|
| 713 |
+
"metadata": {},
|
| 714 |
+
"outputs": [],
|
| 715 |
+
"source": [
|
| 716 |
+
"from zenml.client import Client\n",
|
| 717 |
+
"from IPython.display import display, HTML\n",
|
| 718 |
+
"\n",
|
| 719 |
+
"client = Client()\n",
|
| 720 |
+
"# CHANGE THIS TO THE LATEST RUN ID\n",
|
| 721 |
+
"latest_run = client.get_pipeline_run(\"sentinment_analysis_feature_engineering_pipeline_run_2023_11_21_10_55_56\")\n",
|
| 722 |
+
"html = latest_run.steps[\"evidently_report_step\"].outputs['report_html'].load()\n",
|
| 723 |
+
"display(HTML(html))"
|
| 724 |
+
]
|
| 725 |
+
},
|
| 726 |
+
{
|
| 727 |
+
"cell_type": "markdown",
|
| 728 |
+
"id": "78ab8771-4421-4975-a3d5-12892a56b805",
|
| 729 |
+
"metadata": {},
|
| 730 |
+
"source": [
|
| 731 |
+
"## 💪 Step 2: Train the model with Huggingface Hub as the model registry\n",
|
| 732 |
+
" "
|
| 733 |
+
]
|
| 734 |
+
},
|
| 735 |
+
{
|
| 736 |
+
"cell_type": "markdown",
|
| 737 |
+
"id": "2843efa8-32b6-4b13-ac85-33c99cc94e3e",
|
| 738 |
+
"metadata": {},
|
| 739 |
+
"source": [
|
| 740 |
+
"Once the feature engineering pipeline has run a few times, we have many datasets to choose from. We can feed our desired one into a function that trains the model on the data. Thanks to the ZenML Huggingface integration, this data is loaded directly from the ZenML artifact store.\n",
|
| 741 |
+
"\n",
|
| 742 |
+
"<img src=\"assets/training_pipeline_overview.png\" alt=\"Pipelines Trains\">\n",
|
| 743 |
+
"\n",
|
| 744 |
+
"On the left side, we see our local MLOps stack, which defines our infrastructure and tooling we are using for this particular pipeline. ZenML makes it easy to run on a local stack on your development machine, or switch out the stack to run on a AWS Kubeflow-based stack (if you want to scale up).\n",
|
| 745 |
+
"\n",
|
| 746 |
+
"On the right side is the new kid on the block - the ZenML Model Control Plane. The Model Control Plane is a new feature in ZenML that allows users to have a complete overview of their machine learning models. It allows teams to consolidate all artifacts related to their ML models into one place, and manage its lifecycle easily as you can see from this view from the ZenML Cloud:"
|
| 747 |
+
]
|
| 748 |
+
},
|
| 749 |
+
{
|
| 750 |
+
"cell_type": "code",
|
| 751 |
+
"execution_count": null,
|
| 752 |
+
"id": "4c99b20f-8e3b-4119-86e9-33dd1395470a",
|
| 753 |
+
"metadata": {},
|
| 754 |
+
"outputs": [],
|
| 755 |
+
"source": [
|
| 756 |
+
"pipeline_args[\"config_path\"] = os.path.join(\"configs\", \"trainer_config.yaml\")\n",
|
| 757 |
+
"\n",
|
| 758 |
+
"pipeline_args[\"enable_cache\"] = True\n",
|
| 759 |
+
"\n",
|
| 760 |
+
"run_args_train = {\n",
|
| 761 |
+
" \"num_epochs\": 1,\n",
|
| 762 |
+
" \"train_batch_size\": 64,\n",
|
| 763 |
+
" \"eval_batch_size\": 64,\n",
|
| 764 |
+
" \"learning_rate\": 2e-4,\n",
|
| 765 |
+
" \"weight_decay\": 0.01,\n",
|
| 766 |
+
" \"max_seq_length\": 512,\n",
|
| 767 |
+
"}\n",
|
| 768 |
+
"\n",
|
| 769 |
+
"# Use versioned artifacts from the last step\n",
|
| 770 |
+
"# run_args_train[\"dataset_artifact_id\"] = latest_run.steps['tokenization_step'].output.id\n",
|
| 771 |
+
"# run_args_train[\"tokenizer_artifact_id\"] = latest_run.steps['tokenizer_loader'].output.id\n",
|
| 772 |
+
"\n",
|
| 773 |
+
"# Configure the model\n",
|
| 774 |
+
"pipeline_args[\"model_config\"] = model_config\n",
|
| 775 |
+
"\n",
|
| 776 |
+
"pipeline_args[\n",
|
| 777 |
+
" \"run_name\"\n",
|
| 778 |
+
"] = f\"sentinment_analysis_training_run_{dt.now().strftime('%Y_%m_%d_%H_%M_%S')}\""
|
| 779 |
+
]
|
| 780 |
+
},
|
| 781 |
+
{
|
| 782 |
+
"cell_type": "code",
|
| 783 |
+
"execution_count": null,
|
| 784 |
+
"id": "96592299-0090-4d2a-962e-6ca232c1fb75",
|
| 785 |
+
"metadata": {},
|
| 786 |
+
"outputs": [],
|
| 787 |
+
"source": [
|
| 788 |
+
"sentinment_analysis_training_pipeline.with_options(**pipeline_args)(\n",
|
| 789 |
+
" **run_args_train\n",
|
| 790 |
+
")"
|
| 791 |
+
]
|
| 792 |
+
},
|
| 793 |
+
{
|
| 794 |
+
"cell_type": "code",
|
| 795 |
+
"execution_count": null,
|
| 796 |
+
"id": "e24e29de-6d1b-41da-9ab2-ca2b32f1f540",
|
| 797 |
+
"metadata": {},
|
| 798 |
+
"outputs": [],
|
| 799 |
+
"source": [
|
| 800 |
+
"### Check out a new stack\n",
|
| 801 |
+
"!zenml stack describe hf-sagemaker-airflow"
|
| 802 |
+
]
|
| 803 |
+
},
|
| 804 |
+
{
|
| 805 |
+
"cell_type": "code",
|
| 806 |
+
"execution_count": null,
|
| 807 |
+
"id": "7c9a5bee-8465-4d41-888a-093f1f6a2ef1",
|
| 808 |
+
"metadata": {},
|
| 809 |
+
"outputs": [],
|
| 810 |
+
"source": [
|
| 811 |
+
"### Change the stack\n",
|
| 812 |
+
"!zenml stack set hf-sagemaker-airflow"
|
| 813 |
+
]
|
| 814 |
+
},
|
| 815 |
+
{
|
| 816 |
+
"cell_type": "code",
|
| 817 |
+
"execution_count": null,
|
| 818 |
+
"id": "d3772c50-1c90-4ffc-8394-c9cfca16cc53",
|
| 819 |
+
"metadata": {},
|
| 820 |
+
"outputs": [],
|
| 821 |
+
"source": [
|
| 822 |
+
"sentinment_analysis_training_pipeline.with_options(**pipeline_args)(\n",
|
| 823 |
+
" **run_args_train\n",
|
| 824 |
+
")"
|
| 825 |
+
]
|
| 826 |
+
},
|
| 827 |
+
{
|
| 828 |
+
"cell_type": "markdown",
|
| 829 |
+
"id": "be79f454-a45d-4f5f-aa93-330d52069124",
|
| 830 |
+
"metadata": {},
|
| 831 |
+
"source": [
|
| 832 |
+
"## 🫅 Step 3: Promote the model to production\n"
|
| 833 |
+
]
|
| 834 |
+
},
|
| 835 |
+
{
|
| 836 |
+
"cell_type": "markdown",
|
| 837 |
+
"id": "5a09b432-7a66-473e-bdb6-ffdca730498b",
|
| 838 |
+
"metadata": {},
|
| 839 |
+
"source": [
|
| 840 |
+
"Following training, the automated promotion pipeline evaluates models against predefined metrics, identifying and marking the most performant one as 'Production ready'. This is another common use case for the Model Control Plane; we store the relevant metrics there to access them easily later.\n",
|
| 841 |
+
"\n",
|
| 842 |
+
"<img src=\"assets/promoting_pipeline_overview.png\" alt=\"Pipelines Trains\">"
|
| 843 |
+
]
|
| 844 |
+
},
|
| 845 |
+
{
|
| 846 |
+
"cell_type": "code",
|
| 847 |
+
"execution_count": null,
|
| 848 |
+
"id": "5bac7ae5-70d0-449c-929c-e175c3062f2d",
|
| 849 |
+
"metadata": {},
|
| 850 |
+
"outputs": [],
|
| 851 |
+
"source": [
|
| 852 |
+
"!zenml stack set hf-sagemaker-local"
|
| 853 |
+
]
|
| 854 |
+
},
|
| 855 |
+
{
|
| 856 |
+
"cell_type": "code",
|
| 857 |
+
"execution_count": null,
|
| 858 |
+
"id": "170c9ef6-4e6f-4e50-ac37-e05bef8570ea",
|
| 859 |
+
"metadata": {},
|
| 860 |
+
"outputs": [],
|
| 861 |
+
"source": [
|
| 862 |
+
"run_args_promoting = {}\n",
|
| 863 |
+
"model_config = ModelConfig(name=zenml_model_name)\n",
|
| 864 |
+
"pipeline_args[\"config_path\"] = os.path.join(\"configs\", \"promoting_config.yaml\")\n",
|
| 865 |
+
"\n",
|
| 866 |
+
"pipeline_args[\"model_config\"] = model_config\n",
|
| 867 |
+
"\n",
|
| 868 |
+
"pipeline_args[\n",
|
| 869 |
+
" \"run_name\"\n",
|
| 870 |
+
"] = f\"sentinment_analysis_promoting_pipeline_run_{dt.now().strftime('%Y_%m_%d_%H_%M_%S')}\""
|
| 871 |
+
]
|
| 872 |
+
},
|
| 873 |
+
{
|
| 874 |
+
"cell_type": "code",
|
| 875 |
+
"execution_count": null,
|
| 876 |
+
"id": "e6df11e2-4591-4186-a8f8-243f9c4d1e3d",
|
| 877 |
+
"metadata": {},
|
| 878 |
+
"outputs": [],
|
| 879 |
+
"source": [
|
| 880 |
+
"sentinment_analysis_promote_pipeline.with_options(**pipeline_args)(\n",
|
| 881 |
+
" **run_args_promoting\n",
|
| 882 |
+
")"
|
| 883 |
+
]
|
| 884 |
+
},
|
| 885 |
+
{
|
| 886 |
+
"cell_type": "markdown",
|
| 887 |
+
"id": "6efc4968-35fd-42e3-ba62-d8e1557aa0d6",
|
| 888 |
+
"metadata": {},
|
| 889 |
+
"source": [
|
| 890 |
+
"## 💯 Step 4: Deploy the model to AWS Sagemaker Endpoints\n"
|
| 891 |
+
]
|
| 892 |
+
},
|
| 893 |
+
{
|
| 894 |
+
"cell_type": "markdown",
|
| 895 |
+
"id": "577aff86-bde9-48d4-9b52-209cfed9fd4e",
|
| 896 |
+
"metadata": {},
|
| 897 |
+
"source": [
|
| 898 |
+
"This is the final step to automate the deployment of the slated production model to a Sagemaker endpoint. The deployment pipelines handles the complexities of AWS interactions and ensures that the model, along with its full history and context, is transitioned into a live environment ready for use. Here again we use the Model Control Plane interface to query the Huggingface revision and use that information to push to Huggingface Hub.\n",
|
| 899 |
+
"\n",
|
| 900 |
+
"<img src=\"assets/deploying_pipeline_overview.png\" alt=\"Pipelines Trains\">\n"
|
| 901 |
+
]
|
| 902 |
+
},
|
| 903 |
+
{
|
| 904 |
+
"cell_type": "code",
|
| 905 |
+
"execution_count": null,
|
| 906 |
+
"id": "1513ab5f-de05-4344-9d2c-fedbfbd21ef0",
|
| 907 |
+
"metadata": {},
|
| 908 |
+
"outputs": [],
|
| 909 |
+
"source": [
|
| 910 |
+
"!zenml stack set hf-sagemaker-local"
|
| 911 |
+
]
|
| 912 |
+
},
|
| 913 |
+
{
|
| 914 |
+
"cell_type": "code",
|
| 915 |
+
"execution_count": null,
|
| 916 |
+
"id": "606fdb3c-4eca-4d32-bccb-280743d15528",
|
| 917 |
+
"metadata": {},
|
| 918 |
+
"outputs": [],
|
| 919 |
+
"source": [
|
| 920 |
+
"pipeline_args[\"config_path\"] = os.path.join(\"configs\", \"deploying_config.yaml\")\n",
|
| 921 |
+
"\n",
|
| 922 |
+
"# Deploying pipeline has new ZenML model config\n",
|
| 923 |
+
"model_config = ModelConfig(\n",
|
| 924 |
+
" name=zenml_model_name,\n",
|
| 925 |
+
" version=ModelStages.PRODUCTION,\n",
|
| 926 |
+
")\n",
|
| 927 |
+
"pipeline_args[\"model_config\"] = model_config\n",
|
| 928 |
+
"pipeline_args[\"enable_cache\"] = False\n",
|
| 929 |
+
"run_args_deploying = {}\n",
|
| 930 |
+
"pipeline_args[\n",
|
| 931 |
+
" \"run_name\"\n",
|
| 932 |
+
"] = f\"sentinment_analysis_deploy_pipeline_run_{dt.now().strftime('%Y_%m_%d_%H_%M_%S')}\""
|
| 933 |
+
]
|
| 934 |
+
},
|
| 935 |
+
{
|
| 936 |
+
"cell_type": "code",
|
| 937 |
+
"execution_count": null,
|
| 938 |
+
"id": "87f1f982-ab96-4207-8e7e-e318473587e9",
|
| 939 |
+
"metadata": {},
|
| 940 |
+
"outputs": [],
|
| 941 |
+
"source": [
|
| 942 |
+
"sentinment_analysis_deploy_pipeline.with_options(**pipeline_args)(\n",
|
| 943 |
+
" **run_args_deploying\n",
|
| 944 |
+
")"
|
| 945 |
+
]
|
| 946 |
+
},
|
| 947 |
+
{
|
| 948 |
+
"cell_type": "markdown",
|
| 949 |
+
"id": "594ee4fc-f102-4b99-bdc3-2f1670c87679",
|
| 950 |
+
"metadata": {},
|
| 951 |
+
"source": [
|
| 952 |
+
"ZenML builds upon the straightforward deployment capability of Huggingface models to AWS Sagemaker, and transforms it into a sophisticated, repeatable, and transparent MLOps workflow. It takes charge of the intricate steps necessary for modern ML systems, ensuring that software engineering leads can focus on iteration and innovation rather than operational intricacies.\n",
|
| 953 |
+
"\n",
|
| 954 |
+
"To delve deeper into each stage, refer to the comprehensive guide on GitHub[: zenml-io/zenml-huggingface-sagemak](https://github.com/zenml-io/zenml-huggingface-sagemaker)er. Additionally[, this YouTube playli](https://www.youtube.com/watch?v=Q1EH2H8Akgo&list=PLhNrLW_IWplw6dBbmGcL828-atJMu3CwF)st provides a detailed visual walkthrough of the entire pipeline: Huggingface to Sagemaker ZenML tutorial.\n",
|
| 955 |
+
"\n",
|
| 956 |
+
"Interested in standardizing your MLOps workflows? ZenML Cloud is now available to all - get a managed ZenML server with important features such as RBAC and pipeline trigge[rs. Book a ](https://zenml.io/book-a-demo)demo with us now to learn how you can create your own MLOps pipelines today."
|
| 957 |
+
]
|
| 958 |
+
}
|
| 959 |
+
],
|
| 960 |
+
"metadata": {
|
| 961 |
+
"kernelspec": {
|
| 962 |
+
"display_name": "Python 3 (ipykernel)",
|
| 963 |
+
"language": "python",
|
| 964 |
+
"name": "python3"
|
| 965 |
+
},
|
| 966 |
+
"language_info": {
|
| 967 |
+
"codemirror_mode": {
|
| 968 |
+
"name": "ipython",
|
| 969 |
+
"version": 3
|
| 970 |
+
},
|
| 971 |
+
"file_extension": ".py",
|
| 972 |
+
"mimetype": "text/x-python",
|
| 973 |
+
"name": "python",
|
| 974 |
+
"nbconvert_exporter": "python",
|
| 975 |
+
"pygments_lexer": "ipython3",
|
| 976 |
+
"version": "3.9.13"
|
| 977 |
+
}
|
| 978 |
+
},
|
| 979 |
+
"nbformat": 4,
|
| 980 |
+
"nbformat_minor": 5
|
| 981 |
+
}
|
run.py
ADDED
|
@@ -0,0 +1,173 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# {% include 'templates/license_header' %}
|
| 2 |
+
|
| 3 |
+
import os
|
| 4 |
+
from typing import Optional
|
| 5 |
+
|
| 6 |
+
import click
|
| 7 |
+
from pipelines import (
|
| 8 |
+
feature_engineering,
|
| 9 |
+
inference,
|
| 10 |
+
breast_cancer_training,
|
| 11 |
+
breast_cancer_deployment_pipeline
|
| 12 |
+
)
|
| 13 |
+
from zenml.client import Client
|
| 14 |
+
from zenml.logger import get_logger
|
| 15 |
+
|
| 16 |
+
logger = get_logger(__name__)
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
@click.command(
|
| 20 |
+
help="""
|
| 21 |
+
ZenML Starter project CLI v0.0.1.
|
| 22 |
+
|
| 23 |
+
Run the ZenML starter project with basic options.
|
| 24 |
+
|
| 25 |
+
Examples:
|
| 26 |
+
|
| 27 |
+
\b
|
| 28 |
+
# Run the feature engineering pipeline
|
| 29 |
+
python run.py --feature-pipeline
|
| 30 |
+
|
| 31 |
+
\b
|
| 32 |
+
# Run the training pipeline
|
| 33 |
+
python run.py --training-pipeline
|
| 34 |
+
|
| 35 |
+
\b
|
| 36 |
+
# Run the training pipeline with versioned artifacts
|
| 37 |
+
python run.py --training-pipeline --train-dataset-version-name=1 --test-dataset-version-name=1
|
| 38 |
+
|
| 39 |
+
\b
|
| 40 |
+
# Run the inference pipeline
|
| 41 |
+
python run.py --inference-pipeline
|
| 42 |
+
|
| 43 |
+
"""
|
| 44 |
+
)
|
| 45 |
+
@click.option(
|
| 46 |
+
"--train-dataset-name",
|
| 47 |
+
default="dataset_trn",
|
| 48 |
+
type=click.STRING,
|
| 49 |
+
help="The name of the train dataset produced by feature engineering.",
|
| 50 |
+
)
|
| 51 |
+
@click.option(
|
| 52 |
+
"--train-dataset-version-name",
|
| 53 |
+
default=None,
|
| 54 |
+
type=click.STRING,
|
| 55 |
+
help="Version of the train dataset produced by feature engineering. "
|
| 56 |
+
"If not specified, a new version will be created.",
|
| 57 |
+
)
|
| 58 |
+
@click.option(
|
| 59 |
+
"--test-dataset-name",
|
| 60 |
+
default="dataset_tst",
|
| 61 |
+
type=click.STRING,
|
| 62 |
+
help="The name of the test dataset produced by feature engineering.",
|
| 63 |
+
)
|
| 64 |
+
@click.option(
|
| 65 |
+
"--test-dataset-version-name",
|
| 66 |
+
default=None,
|
| 67 |
+
type=click.STRING,
|
| 68 |
+
help="Version of the test dataset produced by feature engineering. "
|
| 69 |
+
"If not specified, a new version will be created.",
|
| 70 |
+
)
|
| 71 |
+
@click.option(
|
| 72 |
+
"--feature-pipeline",
|
| 73 |
+
is_flag=True,
|
| 74 |
+
default=False,
|
| 75 |
+
help="Whether to run the pipeline that creates the dataset.",
|
| 76 |
+
)
|
| 77 |
+
@click.option(
|
| 78 |
+
"--training-pipeline",
|
| 79 |
+
is_flag=True,
|
| 80 |
+
default=False,
|
| 81 |
+
help="Whether to run the pipeline that trains the model.",
|
| 82 |
+
)
|
| 83 |
+
@click.option(
|
| 84 |
+
"--inference-pipeline",
|
| 85 |
+
is_flag=True,
|
| 86 |
+
default=False,
|
| 87 |
+
help="Whether to run the pipeline that performs inference.",
|
| 88 |
+
)
|
| 89 |
+
@click.option(
|
| 90 |
+
"--deployment-pipeline",
|
| 91 |
+
is_flag=True,
|
| 92 |
+
default=False,
|
| 93 |
+
help="Whether to run the pipeline that deploys the model.",
|
| 94 |
+
)
|
| 95 |
+
def main(
|
| 96 |
+
train_dataset_name: str = "dataset_trn",
|
| 97 |
+
train_dataset_version_name: Optional[str] = None,
|
| 98 |
+
test_dataset_name: str = "dataset_tst",
|
| 99 |
+
test_dataset_version_name: Optional[str] = None,
|
| 100 |
+
feature_pipeline: bool = False,
|
| 101 |
+
training_pipeline: bool = False,
|
| 102 |
+
inference_pipeline: bool = False,
|
| 103 |
+
deployment_pipeline: bool = False,
|
| 104 |
+
):
|
| 105 |
+
"""Main entry point for the pipeline execution.
|
| 106 |
+
|
| 107 |
+
This entrypoint is where everything comes together:
|
| 108 |
+
|
| 109 |
+
* configuring pipeline with the required parameters
|
| 110 |
+
(some of which may come from command line arguments, but most
|
| 111 |
+
of which comes from the YAML config files)
|
| 112 |
+
* launching the pipeline
|
| 113 |
+
"""
|
| 114 |
+
config_folder = os.path.join(
|
| 115 |
+
os.path.dirname(os.path.realpath(__file__)),
|
| 116 |
+
"configs",
|
| 117 |
+
)
|
| 118 |
+
|
| 119 |
+
# Execute Feature Engineering Pipeline
|
| 120 |
+
if feature_pipeline:
|
| 121 |
+
pipeline_args = {}
|
| 122 |
+
pipeline_args["config_path"] = os.path.join(
|
| 123 |
+
config_folder, "feature_engineering.yaml"
|
| 124 |
+
)
|
| 125 |
+
run_args_feature = {}
|
| 126 |
+
feature_engineering.with_options(**pipeline_args)(**run_args_feature)
|
| 127 |
+
logger.info("Feature Engineering pipeline finished successfully!")
|
| 128 |
+
|
| 129 |
+
# Execute Training Pipeline
|
| 130 |
+
if training_pipeline:
|
| 131 |
+
pipeline_args = {}
|
| 132 |
+
pipeline_args["config_path"] = os.path.join(config_folder, "training.yaml")
|
| 133 |
+
|
| 134 |
+
run_args_train = {}
|
| 135 |
+
|
| 136 |
+
# If train_dataset_version_name is specified, use versioned artifacts
|
| 137 |
+
if train_dataset_version_name or test_dataset_version_name:
|
| 138 |
+
# However, both train and test dataset versions must be specified
|
| 139 |
+
assert (
|
| 140 |
+
train_dataset_version_name is not None
|
| 141 |
+
and test_dataset_version_name is not None
|
| 142 |
+
)
|
| 143 |
+
client = Client()
|
| 144 |
+
train_dataset_artifact = client.get_artifact(
|
| 145 |
+
train_dataset_name, train_dataset_version_name
|
| 146 |
+
)
|
| 147 |
+
# If train dataset is specified, test dataset must be specified
|
| 148 |
+
test_dataset_artifact = client.get_artifact(
|
| 149 |
+
test_dataset_name, test_dataset_version_name
|
| 150 |
+
)
|
| 151 |
+
# Use versioned artifacts
|
| 152 |
+
run_args_train["train_dataset_id"] = train_dataset_artifact.id
|
| 153 |
+
run_args_train["test_dataset_id"] = test_dataset_artifact.id
|
| 154 |
+
|
| 155 |
+
breast_cancer_training.with_options(**pipeline_args)(**run_args_train)
|
| 156 |
+
logger.info("Training pipeline finished successfully!")
|
| 157 |
+
|
| 158 |
+
if inference_pipeline:
|
| 159 |
+
pipeline_args = {}
|
| 160 |
+
pipeline_args["config_path"] = os.path.join(config_folder, "inference.yaml")
|
| 161 |
+
run_args_inference = {}
|
| 162 |
+
inference.with_options(**pipeline_args)(**run_args_inference)
|
| 163 |
+
logger.info("Inference pipeline finished successfully!")
|
| 164 |
+
|
| 165 |
+
if deployment_pipeline:
|
| 166 |
+
pipeline_args = {}
|
| 167 |
+
pipeline_args["config_path"] = os.path.join(config_folder, "deployment.yaml")
|
| 168 |
+
run_args_inference = {}
|
| 169 |
+
breast_cancer_deployment_pipeline.with_options(**pipeline_args)(**run_args_inference)
|
| 170 |
+
logger.info("Deployment pipeline finished successfully!")
|
| 171 |
+
|
| 172 |
+
if __name__ == "__main__":
|
| 173 |
+
main()
|
run_stack_showcase.ipynb
ADDED
|
@@ -0,0 +1,347 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
{
|
| 2 |
+
"cells": [
|
| 3 |
+
{
|
| 4 |
+
"cell_type": "code",
|
| 5 |
+
"execution_count": null,
|
| 6 |
+
"id": "b567a1d3-f625-4b98-9852-fcc3f3fe9609",
|
| 7 |
+
"metadata": {},
|
| 8 |
+
"outputs": [],
|
| 9 |
+
"source": [
|
| 10 |
+
"# To start with, we use the default stack\n",
|
| 11 |
+
"#!zenml init\n",
|
| 12 |
+
"\n",
|
| 13 |
+
"# We also need to connect to a remote ZenML Instance\n",
|
| 14 |
+
"#!zenml connect --url https://1cf18d95-zenml.cloudinfra.zenml.io"
|
| 15 |
+
]
|
| 16 |
+
},
|
| 17 |
+
{
|
| 18 |
+
"cell_type": "code",
|
| 19 |
+
"execution_count": null,
|
| 20 |
+
"id": "c53367f1-3951-48c7-9540-21daf818fa5d",
|
| 21 |
+
"metadata": {},
|
| 22 |
+
"outputs": [],
|
| 23 |
+
"source": [
|
| 24 |
+
"# Do the imports at the top\n",
|
| 25 |
+
"\n",
|
| 26 |
+
"import random\n",
|
| 27 |
+
"from zenml import ExternalArtifact, pipeline \n",
|
| 28 |
+
"from zenml.client import Client\n",
|
| 29 |
+
"from zenml.logger import get_logger\n",
|
| 30 |
+
"from uuid import UUID\n",
|
| 31 |
+
"\n",
|
| 32 |
+
"import os\n",
|
| 33 |
+
"from typing import Optional, List\n",
|
| 34 |
+
"\n",
|
| 35 |
+
"from zenml import pipeline\n",
|
| 36 |
+
"from zenml.model.model_version import ModelVersion\n",
|
| 37 |
+
"\n",
|
| 38 |
+
"from pipelines import feature_engineering\n",
|
| 39 |
+
"\n",
|
| 40 |
+
"from steps import (\n",
|
| 41 |
+
" data_loader,\n",
|
| 42 |
+
" data_preprocessor,\n",
|
| 43 |
+
" data_splitter,\n",
|
| 44 |
+
" model_evaluator,\n",
|
| 45 |
+
" model_trainer,\n",
|
| 46 |
+
" inference_predict,\n",
|
| 47 |
+
" inference_preprocessor\n",
|
| 48 |
+
")\n",
|
| 49 |
+
"\n",
|
| 50 |
+
"logger = get_logger(__name__)\n",
|
| 51 |
+
"\n",
|
| 52 |
+
"client = Client()\n",
|
| 53 |
+
"client.activate_stack(\"local-mlflow-stack\")"
|
| 54 |
+
]
|
| 55 |
+
},
|
| 56 |
+
{
|
| 57 |
+
"cell_type": "markdown",
|
| 58 |
+
"id": "ab87746e-b804-4fab-88f6-d4967048cb45",
|
| 59 |
+
"metadata": {},
|
| 60 |
+
"source": [
|
| 61 |
+
"# Start local with a simple training pipeline\n",
|
| 62 |
+
"\n",
|
| 63 |
+
"Below you can see what the pipeline looks like. We will start by running this locally on the default-stack. This means the data between the steps is stored locally and the compute is also local."
|
| 64 |
+
]
|
| 65 |
+
},
|
| 66 |
+
{
|
| 67 |
+
"cell_type": "markdown",
|
| 68 |
+
"id": "33872b19-7329-4f5e-9a1e-cfc1fe9d560d",
|
| 69 |
+
"metadata": {
|
| 70 |
+
"jp-MarkdownHeadingCollapsed": true
|
| 71 |
+
},
|
| 72 |
+
"source": [
|
| 73 |
+
"<img src=\"_assets/default_stack.png\" alt=\"Drawing\" style=\"width: 800px;\"/>"
|
| 74 |
+
]
|
| 75 |
+
},
|
| 76 |
+
{
|
| 77 |
+
"cell_type": "code",
|
| 78 |
+
"execution_count": null,
|
| 79 |
+
"id": "06625571-b281-4820-a7eb-3a085ba2e572",
|
| 80 |
+
"metadata": {},
|
| 81 |
+
"outputs": [],
|
| 82 |
+
"source": [
|
| 83 |
+
"import pandas as pd\n",
|
| 84 |
+
"from sklearn.datasets import load_breast_cancer\n",
|
| 85 |
+
"from zenml import step\n",
|
| 86 |
+
"from zenml.logger import get_logger\n",
|
| 87 |
+
"\n",
|
| 88 |
+
"logger = get_logger(__name__)\n",
|
| 89 |
+
"\n",
|
| 90 |
+
"# Here is what one of the steps in the pipeline looks like. Simple python function that just needs the `@step` decorator.\n",
|
| 91 |
+
"\n",
|
| 92 |
+
"@step\n",
|
| 93 |
+
"def data_loader() -> pd.DataFrame:\n",
|
| 94 |
+
" \"\"\"Dataset reader step.\"\"\"\n",
|
| 95 |
+
" dataset = load_breast_cancer(as_frame=True)\n",
|
| 96 |
+
" inference_size = int(len(dataset.target) * 0.05)\n",
|
| 97 |
+
" dataset: pd.DataFrame = dataset.frame\n",
|
| 98 |
+
" dataset.reset_index(drop=True, inplace=True)\n",
|
| 99 |
+
" logger.info(f\"Dataset with {len(dataset)} records loaded!\")\n",
|
| 100 |
+
"\n",
|
| 101 |
+
" return dataset\n"
|
| 102 |
+
]
|
| 103 |
+
},
|
| 104 |
+
{
|
| 105 |
+
"cell_type": "code",
|
| 106 |
+
"execution_count": null,
|
| 107 |
+
"id": "754a3069-9d13-4869-be64-a641071800cc",
|
| 108 |
+
"metadata": {},
|
| 109 |
+
"outputs": [],
|
| 110 |
+
"source": [
|
| 111 |
+
"# Here's an example of what this function returns\n",
|
| 112 |
+
"\n",
|
| 113 |
+
"data_loader()"
|
| 114 |
+
]
|
| 115 |
+
},
|
| 116 |
+
{
|
| 117 |
+
"cell_type": "code",
|
| 118 |
+
"execution_count": null,
|
| 119 |
+
"id": "8aa300f1-48df-4e62-87eb-0e2fc5735da8",
|
| 120 |
+
"metadata": {},
|
| 121 |
+
"outputs": [],
|
| 122 |
+
"source": [
|
| 123 |
+
"from zenml import pipeline\n",
|
| 124 |
+
"\n",
|
| 125 |
+
"@pipeline\n",
|
| 126 |
+
"def breast_cancer_training(\n",
|
| 127 |
+
" train_dataset_id: Optional[UUID] = None,\n",
|
| 128 |
+
" test_dataset_id: Optional[UUID] = None,\n",
|
| 129 |
+
" min_train_accuracy: float = 0.0,\n",
|
| 130 |
+
" min_test_accuracy: float = 0.0,\n",
|
| 131 |
+
"):\n",
|
| 132 |
+
" \"\"\"Model training pipeline.\"\"\"\n",
|
| 133 |
+
" # Execute Feature Engineering Pipeline\n",
|
| 134 |
+
" dataset_trn, dataset_tst = feature_engineering()\n",
|
| 135 |
+
"\n",
|
| 136 |
+
" model = model_trainer(\n",
|
| 137 |
+
" dataset_trn=dataset_trn,\n",
|
| 138 |
+
" )\n",
|
| 139 |
+
"\n",
|
| 140 |
+
" model_evaluator(\n",
|
| 141 |
+
" model=model,\n",
|
| 142 |
+
" dataset_trn=dataset_trn,\n",
|
| 143 |
+
" dataset_tst=dataset_tst,\n",
|
| 144 |
+
" min_train_accuracy=min_train_accuracy,\n",
|
| 145 |
+
" min_test_accuracy=min_test_accuracy,\n",
|
| 146 |
+
" )\n"
|
| 147 |
+
]
|
| 148 |
+
},
|
| 149 |
+
{
|
| 150 |
+
"cell_type": "code",
|
| 151 |
+
"execution_count": null,
|
| 152 |
+
"id": "d55342bf-33c5-4646-b1ce-e599a99cf568",
|
| 153 |
+
"metadata": {},
|
| 154 |
+
"outputs": [],
|
| 155 |
+
"source": [
|
| 156 |
+
"model_version = ModelVersion(\n",
|
| 157 |
+
" name=\"breast_cancer_classifier_model\",\n",
|
| 158 |
+
" description=\"Classification of Breast Cancer Dataset.\",\n",
|
| 159 |
+
" delete_new_version_on_failure=True,\n",
|
| 160 |
+
" tags=[\"classification\", \"sklearn\"],\n",
|
| 161 |
+
")\n",
|
| 162 |
+
"\n",
|
| 163 |
+
"pipeline_args = {\n",
|
| 164 |
+
" \"enable_cache\": True, \n",
|
| 165 |
+
" \"model_version\": model_version\n",
|
| 166 |
+
"}\n",
|
| 167 |
+
"\n",
|
| 168 |
+
"# Model Version config\n",
|
| 169 |
+
"fe_t_configured = breast_cancer_training.with_options(**pipeline_args)"
|
| 170 |
+
]
|
| 171 |
+
},
|
| 172 |
+
{
|
| 173 |
+
"cell_type": "code",
|
| 174 |
+
"execution_count": null,
|
| 175 |
+
"id": "f5f4aed8-7d87-4e07-a25c-345d327ad636",
|
| 176 |
+
"metadata": {},
|
| 177 |
+
"outputs": [],
|
| 178 |
+
"source": [
|
| 179 |
+
"fe_t_configured()"
|
| 180 |
+
]
|
| 181 |
+
},
|
| 182 |
+
{
|
| 183 |
+
"cell_type": "markdown",
|
| 184 |
+
"id": "c3e6dc42-21b8-4b3c-90ec-d6e6d541907f",
|
| 185 |
+
"metadata": {},
|
| 186 |
+
"source": [
|
| 187 |
+
"# Let's outsource some compute to Sagemaker!"
|
| 188 |
+
]
|
| 189 |
+
},
|
| 190 |
+
{
|
| 191 |
+
"cell_type": "markdown",
|
| 192 |
+
"id": "14a840b1-288d-4713-98f4-bbe8d6e06140",
|
| 193 |
+
"metadata": {},
|
| 194 |
+
"source": [
|
| 195 |
+
"Let's farm some compute to AWS with a training job with a certain number of CPUs and Memory. This can easily be done without and changes to the actual implementation of the pipeline. "
|
| 196 |
+
]
|
| 197 |
+
},
|
| 198 |
+
{
|
| 199 |
+
"cell_type": "markdown",
|
| 200 |
+
"id": "fa9308fb-3556-472c-8fc7-7f2f88d1c455",
|
| 201 |
+
"metadata": {},
|
| 202 |
+
"source": [
|
| 203 |
+
"\n",
|
| 204 |
+
"\n",
|
| 205 |
+
"\n",
|
| 206 |
+
"\n",
|
| 207 |
+
"\n",
|
| 208 |
+
"\n",
|
| 209 |
+
"\n",
|
| 210 |
+
"\n",
|
| 211 |
+
"\n",
|
| 212 |
+
"\n",
|
| 213 |
+
"\n",
|
| 214 |
+
"\n",
|
| 215 |
+
"\n",
|
| 216 |
+
"<img src=\"_assets/local_sagmaker_so_stack.png\" alt=\"Drawing\" style=\"width: 800px;\"/>"
|
| 217 |
+
]
|
| 218 |
+
},
|
| 219 |
+
{
|
| 220 |
+
"cell_type": "code",
|
| 221 |
+
"execution_count": null,
|
| 222 |
+
"id": "48be8f60-9fbe-4d19-92e4-d9cd8289dbf7",
|
| 223 |
+
"metadata": {
|
| 224 |
+
"scrolled": true
|
| 225 |
+
},
|
| 226 |
+
"outputs": [],
|
| 227 |
+
"source": [
|
| 228 |
+
"# This pip installs the requirements locally\n",
|
| 229 |
+
"!zenml integration install aws s3 mlflow -y"
|
| 230 |
+
]
|
| 231 |
+
},
|
| 232 |
+
{
|
| 233 |
+
"cell_type": "code",
|
| 234 |
+
"execution_count": null,
|
| 235 |
+
"id": "4cb26018-aa7d-497d-a0e2-855d3becb70d",
|
| 236 |
+
"metadata": {},
|
| 237 |
+
"outputs": [],
|
| 238 |
+
"source": [
|
| 239 |
+
"client.activate_stack(\"local-sagemaker-step-operator-stack\")"
|
| 240 |
+
]
|
| 241 |
+
},
|
| 242 |
+
{
|
| 243 |
+
"cell_type": "code",
|
| 244 |
+
"execution_count": null,
|
| 245 |
+
"id": "5683a1c9-f5c1-4ba1-ad7c-1e427fd265df",
|
| 246 |
+
"metadata": {},
|
| 247 |
+
"outputs": [],
|
| 248 |
+
"source": [
|
| 249 |
+
"from zenml.config import DockerSettings\n",
|
| 250 |
+
"\n",
|
| 251 |
+
"# The actual code will stay the same, all that needs to be done is some configuration\n",
|
| 252 |
+
"step_args = {}\n",
|
| 253 |
+
"\n",
|
| 254 |
+
"# We configure which step operator should be used\n",
|
| 255 |
+
"step_args[\"step_operator\"] = \"sagemaker-eu\"\n",
|
| 256 |
+
"\n",
|
| 257 |
+
"# M5 Large is what we need for this big data!\n",
|
| 258 |
+
"step_args[\"settings\"] = {\"step_operator.sagemaker\": {\"estimator_args\": {\"instance_type\" : \"ml.m5.large\"}}}\n",
|
| 259 |
+
"\n",
|
| 260 |
+
"# Update the step. We could also do this in YAML\n",
|
| 261 |
+
"model_trainer = model_trainer.with_options(**step_args)\n",
|
| 262 |
+
"\n",
|
| 263 |
+
"docker_settings = DockerSettings(\n",
|
| 264 |
+
" requirements=[\n",
|
| 265 |
+
" \"pyarrow\",\n",
|
| 266 |
+
" \"scikit-learn==1.1.1\"\n",
|
| 267 |
+
" ],\n",
|
| 268 |
+
")\n",
|
| 269 |
+
"\n",
|
| 270 |
+
"pipeline_args = {\n",
|
| 271 |
+
" \"enable_cache\": True, \n",
|
| 272 |
+
" \"model_version\": model_version,\n",
|
| 273 |
+
" \"settings\": {\"docker\": docker_settings}\n",
|
| 274 |
+
"}\n",
|
| 275 |
+
"\n",
|
| 276 |
+
"fe_t_configured = breast_cancer_training.with_options(**pipeline_args)"
|
| 277 |
+
]
|
| 278 |
+
},
|
| 279 |
+
{
|
| 280 |
+
"cell_type": "code",
|
| 281 |
+
"execution_count": null,
|
| 282 |
+
"id": "85179f52-68f0-4c8d-9808-6b080bec72c3",
|
| 283 |
+
"metadata": {
|
| 284 |
+
"scrolled": true
|
| 285 |
+
},
|
| 286 |
+
"outputs": [],
|
| 287 |
+
"source": [
|
| 288 |
+
"# Lets run the pipeline\n",
|
| 289 |
+
"fe_t_configured()"
|
| 290 |
+
]
|
| 291 |
+
},
|
| 292 |
+
{
|
| 293 |
+
"cell_type": "markdown",
|
| 294 |
+
"id": "0841f93b-9eb5-4af6-bba7-cec167024ccf",
|
| 295 |
+
"metadata": {},
|
| 296 |
+
"source": [
|
| 297 |
+
"# Switch to full Sagemaker Stack\n",
|
| 298 |
+
"\n",
|
| 299 |
+
"Just one command will allow you to switch the full code execution over to sagemaker. No Sagemaker domain knowledge necessary. No setup of VMs or Kubernetes clusters necessary. No maintenance of any infrastructure either.\n",
|
| 300 |
+
"\n",
|
| 301 |
+
"\n"
|
| 302 |
+
]
|
| 303 |
+
},
|
| 304 |
+
{
|
| 305 |
+
"cell_type": "code",
|
| 306 |
+
"execution_count": null,
|
| 307 |
+
"id": "d8e33484-3377-4f0e-83fa-87d7c0ca4d72",
|
| 308 |
+
"metadata": {},
|
| 309 |
+
"outputs": [],
|
| 310 |
+
"source": [
|
| 311 |
+
"# Finally, this is all that needs to be done to fully switch the code to be run fully on sagemaker\n",
|
| 312 |
+
"client.activate_stack(\"sagemaker-stack\")"
|
| 313 |
+
]
|
| 314 |
+
},
|
| 315 |
+
{
|
| 316 |
+
"cell_type": "code",
|
| 317 |
+
"execution_count": null,
|
| 318 |
+
"id": "a03c95e9-df2e-446c-8d61-9cc37ad8a46a",
|
| 319 |
+
"metadata": {},
|
| 320 |
+
"outputs": [],
|
| 321 |
+
"source": [
|
| 322 |
+
"fe_t_configured()"
|
| 323 |
+
]
|
| 324 |
+
}
|
| 325 |
+
],
|
| 326 |
+
"metadata": {
|
| 327 |
+
"kernelspec": {
|
| 328 |
+
"display_name": "Python 3 (ipykernel)",
|
| 329 |
+
"language": "python",
|
| 330 |
+
"name": "python3"
|
| 331 |
+
},
|
| 332 |
+
"language_info": {
|
| 333 |
+
"codemirror_mode": {
|
| 334 |
+
"name": "ipython",
|
| 335 |
+
"version": 3
|
| 336 |
+
},
|
| 337 |
+
"file_extension": ".py",
|
| 338 |
+
"mimetype": "text/x-python",
|
| 339 |
+
"name": "python",
|
| 340 |
+
"nbconvert_exporter": "python",
|
| 341 |
+
"pygments_lexer": "ipython3",
|
| 342 |
+
"version": "3.8.10"
|
| 343 |
+
}
|
| 344 |
+
},
|
| 345 |
+
"nbformat": 4,
|
| 346 |
+
"nbformat_minor": 5
|
| 347 |
+
}
|
steps/__init__.py
ADDED
|
@@ -0,0 +1,29 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# {% include 'template/license_header' %}
|
| 2 |
+
|
| 3 |
+
from .data_loader import (
|
| 4 |
+
data_loader,
|
| 5 |
+
)
|
| 6 |
+
from .data_preprocessor import (
|
| 7 |
+
data_preprocessor,
|
| 8 |
+
)
|
| 9 |
+
from .data_splitter import (
|
| 10 |
+
data_splitter,
|
| 11 |
+
)
|
| 12 |
+
from .inference_predict import (
|
| 13 |
+
inference_predict,
|
| 14 |
+
)
|
| 15 |
+
from .inference_preprocessor import (
|
| 16 |
+
inference_preprocessor,
|
| 17 |
+
)
|
| 18 |
+
from .model_evaluator import (
|
| 19 |
+
model_evaluator,
|
| 20 |
+
)
|
| 21 |
+
from .model_trainer import (
|
| 22 |
+
model_trainer,
|
| 23 |
+
)
|
| 24 |
+
from .model_promoter import (
|
| 25 |
+
model_promoter,
|
| 26 |
+
)
|
| 27 |
+
from .deploy_to_huggingface import (
|
| 28 |
+
deploy_to_huggingface,
|
| 29 |
+
)
|
steps/__pycache__/__init__.cpython-38.pyc
ADDED
|
Binary file (521 Bytes). View file
|
|
|
steps/__pycache__/data_loader.cpython-38.pyc
ADDED
|
Binary file (1.91 kB). View file
|
|
|
steps/__pycache__/data_preprocessor.cpython-38.pyc
ADDED
|
Binary file (4.23 kB). View file
|
|
|
steps/__pycache__/data_splitter.cpython-38.pyc
ADDED
|
Binary file (1.52 kB). View file
|
|
|
steps/__pycache__/deploy_to_huggingface.cpython-38.pyc
ADDED
|
Binary file (1.57 kB). View file
|
|
|
steps/__pycache__/inference_predict.cpython-38.pyc
ADDED
|
Binary file (1.32 kB). View file
|
|
|
steps/__pycache__/inference_preprocessor.cpython-38.pyc
ADDED
|
Binary file (1.26 kB). View file
|
|
|
steps/__pycache__/model_evaluator.cpython-38.pyc
ADDED
|
Binary file (3.49 kB). View file
|
|
|
steps/__pycache__/model_promoter.cpython-38.pyc
ADDED
|
Binary file (1.43 kB). View file
|
|
|
steps/__pycache__/model_trainer.cpython-38.pyc
ADDED
|
Binary file (1.58 kB). View file
|
|
|
steps/data_loader.py
ADDED
|
@@ -0,0 +1,53 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# {% include 'template/license_header' %}
|
| 2 |
+
|
| 3 |
+
import pandas as pd
|
| 4 |
+
from sklearn.datasets import load_breast_cancer
|
| 5 |
+
from typing_extensions import Annotated
|
| 6 |
+
from zenml import log_artifact_metadata, step
|
| 7 |
+
from zenml.logger import get_logger
|
| 8 |
+
|
| 9 |
+
logger = get_logger(__name__)
|
| 10 |
+
|
| 11 |
+
|
| 12 |
+
@step
|
| 13 |
+
def data_loader(
|
| 14 |
+
random_state: int, is_inference: bool = False, target: str = "target"
|
| 15 |
+
) -> Annotated[pd.DataFrame, "dataset"]:
|
| 16 |
+
"""Dataset reader step.
|
| 17 |
+
|
| 18 |
+
This is an example of a dataset reader step that load Breast Cancer dataset.
|
| 19 |
+
|
| 20 |
+
This step is parameterized, which allows you to configure the step
|
| 21 |
+
independently of the step code, before running it in a pipeline.
|
| 22 |
+
In this example, the step can be configured with number of rows and logic
|
| 23 |
+
to drop target column or not. See the documentation for more information:
|
| 24 |
+
|
| 25 |
+
https://docs.zenml.io/user-guide/advanced-guide/configure-steps-pipelines
|
| 26 |
+
|
| 27 |
+
Args:
|
| 28 |
+
is_inference: If `True` subset will be returned and target column
|
| 29 |
+
will be removed from dataset.
|
| 30 |
+
random_state: Random state for sampling
|
| 31 |
+
target: Name of target columns in dataset.
|
| 32 |
+
|
| 33 |
+
Returns:
|
| 34 |
+
The dataset artifact as Pandas DataFrame and name of target column.
|
| 35 |
+
"""
|
| 36 |
+
### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###
|
| 37 |
+
dataset = load_breast_cancer(as_frame=True)
|
| 38 |
+
inference_size = int(len(dataset.target) * 0.05)
|
| 39 |
+
dataset: pd.DataFrame = dataset.frame
|
| 40 |
+
inference_subset = dataset.sample(inference_size, random_state=random_state)
|
| 41 |
+
if is_inference:
|
| 42 |
+
dataset = inference_subset
|
| 43 |
+
dataset.drop(columns=target, inplace=True)
|
| 44 |
+
else:
|
| 45 |
+
dataset.drop(inference_subset.index, inplace=True)
|
| 46 |
+
dataset.reset_index(drop=True, inplace=True)
|
| 47 |
+
logger.info(f"Dataset with {len(dataset)} records loaded!")
|
| 48 |
+
|
| 49 |
+
# Recording metadata for this dataset
|
| 50 |
+
log_artifact_metadata(metadata={"random_state": random_state, target: target})
|
| 51 |
+
|
| 52 |
+
### YOUR CODE ENDS HERE ###
|
| 53 |
+
return dataset
|
steps/data_preprocessor.py
ADDED
|
@@ -0,0 +1,115 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# {% include 'template/license_header' %}
|
| 2 |
+
|
| 3 |
+
from typing import Union
|
| 4 |
+
import pandas as pd
|
| 5 |
+
from typing import List, Optional, Tuple
|
| 6 |
+
|
| 7 |
+
import pandas as pd
|
| 8 |
+
from sklearn.pipeline import Pipeline
|
| 9 |
+
from sklearn.preprocessing import MinMaxScaler
|
| 10 |
+
from typing_extensions import Annotated
|
| 11 |
+
from zenml import log_artifact_metadata, step
|
| 12 |
+
|
| 13 |
+
|
| 14 |
+
class NADropper:
|
| 15 |
+
"""Support class to drop NA values in sklearn Pipeline."""
|
| 16 |
+
|
| 17 |
+
def fit(self, *args, **kwargs):
|
| 18 |
+
return self
|
| 19 |
+
|
| 20 |
+
def transform(self, X: Union[pd.DataFrame, pd.Series]):
|
| 21 |
+
return X.dropna()
|
| 22 |
+
|
| 23 |
+
|
| 24 |
+
class ColumnsDropper:
|
| 25 |
+
"""Support class to drop specific columns in sklearn Pipeline."""
|
| 26 |
+
|
| 27 |
+
def __init__(self, columns):
|
| 28 |
+
self.columns = columns
|
| 29 |
+
|
| 30 |
+
def fit(self, *args, **kwargs):
|
| 31 |
+
return self
|
| 32 |
+
|
| 33 |
+
def transform(self, X: Union[pd.DataFrame, pd.Series]):
|
| 34 |
+
return X.drop(columns=self.columns)
|
| 35 |
+
|
| 36 |
+
|
| 37 |
+
class DataFrameCaster:
|
| 38 |
+
"""Support class to cast type back to pd.DataFrame in sklearn Pipeline."""
|
| 39 |
+
|
| 40 |
+
def __init__(self, columns):
|
| 41 |
+
self.columns = columns
|
| 42 |
+
|
| 43 |
+
def fit(self, *args, **kwargs):
|
| 44 |
+
return self
|
| 45 |
+
|
| 46 |
+
def transform(self, X):
|
| 47 |
+
return pd.DataFrame(X, columns=self.columns)
|
| 48 |
+
|
| 49 |
+
|
| 50 |
+
@step
|
| 51 |
+
def data_preprocessor(
|
| 52 |
+
dataset_trn: pd.DataFrame,
|
| 53 |
+
dataset_tst: pd.DataFrame,
|
| 54 |
+
drop_na: Optional[bool] = None,
|
| 55 |
+
normalize: Optional[bool] = None,
|
| 56 |
+
drop_columns: Optional[List[str]] = None,
|
| 57 |
+
target: Optional[str] = "target",
|
| 58 |
+
) -> Tuple[
|
| 59 |
+
Annotated[pd.DataFrame, "dataset_trn"],
|
| 60 |
+
Annotated[pd.DataFrame, "dataset_tst"],
|
| 61 |
+
Annotated[Pipeline, "preprocess_pipeline"],
|
| 62 |
+
]:
|
| 63 |
+
"""Data preprocessor step.
|
| 64 |
+
|
| 65 |
+
This is an example of a data processor step that prepares the data so that
|
| 66 |
+
it is suitable for model training. It takes in a dataset as an input step
|
| 67 |
+
artifact and performs any necessary preprocessing steps like cleaning,
|
| 68 |
+
feature engineering, feature selection, etc. It then returns the processed
|
| 69 |
+
dataset as an step output artifact.
|
| 70 |
+
|
| 71 |
+
This step is parameterized, which allows you to configure the step
|
| 72 |
+
independently of the step code, before running it in a pipeline.
|
| 73 |
+
In this example, the step can be configured to drop NA values, drop some
|
| 74 |
+
columns and normalize numerical columns. See the documentation for more
|
| 75 |
+
information:
|
| 76 |
+
|
| 77 |
+
https://docs.zenml.io/user-guide/advanced-guide/configure-steps-pipelines
|
| 78 |
+
|
| 79 |
+
Args:
|
| 80 |
+
dataset_trn: The train dataset.
|
| 81 |
+
dataset_tst: The test dataset.
|
| 82 |
+
drop_na: If `True` all NA rows will be dropped.
|
| 83 |
+
normalize: If `True` all numeric fields will be normalized.
|
| 84 |
+
drop_columns: List of column names to drop.
|
| 85 |
+
|
| 86 |
+
Returns:
|
| 87 |
+
The processed datasets (dataset_trn, dataset_tst) and fitted `Pipeline` object.
|
| 88 |
+
"""
|
| 89 |
+
### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###
|
| 90 |
+
# We use the sklearn pipeline to chain together multiple preprocessing steps
|
| 91 |
+
preprocess_pipeline = Pipeline([("passthrough", "passthrough")])
|
| 92 |
+
if drop_na:
|
| 93 |
+
preprocess_pipeline.steps.append(("drop_na", NADropper()))
|
| 94 |
+
if drop_columns:
|
| 95 |
+
# Drop columns
|
| 96 |
+
preprocess_pipeline.steps.append(("drop_columns", ColumnsDropper(drop_columns)))
|
| 97 |
+
if normalize:
|
| 98 |
+
# Normalize the data
|
| 99 |
+
preprocess_pipeline.steps.append(("normalize", MinMaxScaler()))
|
| 100 |
+
preprocess_pipeline.steps.append(("cast", DataFrameCaster(dataset_trn.columns)))
|
| 101 |
+
dataset_trn = preprocess_pipeline.fit_transform(dataset_trn)
|
| 102 |
+
dataset_tst = preprocess_pipeline.transform(dataset_tst)
|
| 103 |
+
|
| 104 |
+
# Log metadata of target to both datasets
|
| 105 |
+
log_artifact_metadata(
|
| 106 |
+
artifact_name="dataset_trn",
|
| 107 |
+
metadata={"target": target},
|
| 108 |
+
)
|
| 109 |
+
log_artifact_metadata(
|
| 110 |
+
artifact_name="dataset_tst",
|
| 111 |
+
metadata={"target": target},
|
| 112 |
+
)
|
| 113 |
+
|
| 114 |
+
### YOUR CODE ENDS HERE ###
|
| 115 |
+
return dataset_trn, dataset_tst, preprocess_pipeline
|
steps/data_splitter.py
ADDED
|
@@ -0,0 +1,47 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# {% include 'template/license_header' %}
|
| 2 |
+
|
| 3 |
+
from typing import Tuple
|
| 4 |
+
|
| 5 |
+
import pandas as pd
|
| 6 |
+
from sklearn.model_selection import train_test_split
|
| 7 |
+
from typing_extensions import Annotated
|
| 8 |
+
from zenml import step
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
@step
|
| 12 |
+
def data_splitter(
|
| 13 |
+
dataset: pd.DataFrame, test_size: float = 0.2
|
| 14 |
+
) -> Tuple[
|
| 15 |
+
Annotated[pd.DataFrame, "raw_dataset_trn"],
|
| 16 |
+
Annotated[pd.DataFrame, "raw_dataset_tst"],
|
| 17 |
+
]:
|
| 18 |
+
"""Dataset splitter step.
|
| 19 |
+
|
| 20 |
+
This is an example of a dataset splitter step that splits the data
|
| 21 |
+
into train and test set before passing it to ML model.
|
| 22 |
+
|
| 23 |
+
This step is parameterized, which allows you to configure the step
|
| 24 |
+
independently of the step code, before running it in a pipeline.
|
| 25 |
+
In this example, the step can be configured to use different test
|
| 26 |
+
set sizes. See the documentation for more information:
|
| 27 |
+
|
| 28 |
+
https://docs.zenml.io/user-guide/advanced-guide/configure-steps-pipelines
|
| 29 |
+
|
| 30 |
+
Args:
|
| 31 |
+
dataset: Dataset read from source.
|
| 32 |
+
test_size: 0.0..1.0 defining portion of test set.
|
| 33 |
+
|
| 34 |
+
Returns:
|
| 35 |
+
The split dataset: dataset_trn, dataset_tst.
|
| 36 |
+
"""
|
| 37 |
+
### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###
|
| 38 |
+
dataset_trn, dataset_tst = train_test_split(
|
| 39 |
+
dataset,
|
| 40 |
+
test_size=test_size,
|
| 41 |
+
random_state=42,
|
| 42 |
+
shuffle=True,
|
| 43 |
+
)
|
| 44 |
+
dataset_trn = pd.DataFrame(dataset_trn, columns=dataset.columns)
|
| 45 |
+
dataset_tst = pd.DataFrame(dataset_tst, columns=dataset.columns)
|
| 46 |
+
### YOUR CODE ENDS HERE ###
|
| 47 |
+
return dataset_trn, dataset_tst
|
steps/deploy_to_huggingface.py
ADDED
|
@@ -0,0 +1,58 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Apache Software License 2.0
|
| 2 |
+
#
|
| 3 |
+
# Copyright (c) ZenML GmbH 2023. All rights reserved.
|
| 4 |
+
#
|
| 5 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 6 |
+
# you may not use this file except in compliance with the License.
|
| 7 |
+
# You may obtain a copy of the License at
|
| 8 |
+
#
|
| 9 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 10 |
+
#
|
| 11 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 12 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 13 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 14 |
+
# See the License for the specific language governing permissions and
|
| 15 |
+
# limitations under the License.
|
| 16 |
+
#
|
| 17 |
+
|
| 18 |
+
import os
|
| 19 |
+
from typing import Optional, List
|
| 20 |
+
from huggingface_hub import create_branch, login, HfApi
|
| 21 |
+
|
| 22 |
+
from zenml import step
|
| 23 |
+
from zenml.client import Client
|
| 24 |
+
from zenml.logger import get_logger
|
| 25 |
+
|
| 26 |
+
# Initialize logger
|
| 27 |
+
logger = get_logger(__name__)
|
| 28 |
+
|
| 29 |
+
|
| 30 |
+
@step
|
| 31 |
+
def deploy_to_huggingface(
|
| 32 |
+
repo_name: str,
|
| 33 |
+
):
|
| 34 |
+
"""
|
| 35 |
+
This step deploy the model to huggingface.
|
| 36 |
+
|
| 37 |
+
Args:
|
| 38 |
+
repo_name: The name of the repo to create/use on huggingface.
|
| 39 |
+
"""
|
| 40 |
+
### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###
|
| 41 |
+
secret = Client().get_secret("huggingface_creds")
|
| 42 |
+
assert secret, "No secret found with name 'huggingface_creds'. Please create one that includes your `username` and `token`."
|
| 43 |
+
token = secret.secret_values["token"]
|
| 44 |
+
api = HfApi(token=token)
|
| 45 |
+
hf_repo = api.create_repo(repo_id=repo_name, repo_type="space", space_sdk="gradio", exist_ok=True)
|
| 46 |
+
zenml_repo_root = Client().root
|
| 47 |
+
if not zenml_repo_root:
|
| 48 |
+
logger.warning(
|
| 49 |
+
"You're running the `deploy_to_huggingface` step outside of a ZenML repo. "
|
| 50 |
+
"Since the deployment step to huggingface is all about pushing the repo to huggingface, "
|
| 51 |
+
"this step will not work outside of a ZenML repo where the gradio folder is present."
|
| 52 |
+
)
|
| 53 |
+
raise
|
| 54 |
+
space = api.upload_folder(
|
| 55 |
+
folder_path=zenml_repo_root, repo_id=hf_repo.repo_id, repo_type="space",
|
| 56 |
+
)
|
| 57 |
+
logger.info(f"Space created: {space}")
|
| 58 |
+
### YOUR CODE ENDS HERE ###
|
steps/inference_predict.py
ADDED
|
@@ -0,0 +1,59 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Apache Software License 2.0
|
| 2 |
+
#
|
| 3 |
+
# Copyright (c) ZenML GmbH 2023. All rights reserved.
|
| 4 |
+
#
|
| 5 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 6 |
+
# you may not use this file except in compliance with the License.
|
| 7 |
+
# You may obtain a copy of the License at
|
| 8 |
+
#
|
| 9 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 10 |
+
#
|
| 11 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 12 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 13 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 14 |
+
# See the License for the specific language governing permissions and
|
| 15 |
+
# limitations under the License.
|
| 16 |
+
#
|
| 17 |
+
|
| 18 |
+
|
| 19 |
+
import pandas as pd
|
| 20 |
+
from typing_extensions import Annotated
|
| 21 |
+
from zenml import get_step_context, step
|
| 22 |
+
from zenml.logger import get_logger
|
| 23 |
+
|
| 24 |
+
logger = get_logger(__name__)
|
| 25 |
+
|
| 26 |
+
|
| 27 |
+
@step
|
| 28 |
+
def inference_predict(
|
| 29 |
+
dataset_inf: pd.DataFrame,
|
| 30 |
+
) -> Annotated[pd.Series, "predictions"]:
|
| 31 |
+
"""Predictions step.
|
| 32 |
+
|
| 33 |
+
This is an example of a predictions step that takes the data in and returns
|
| 34 |
+
predicted values.
|
| 35 |
+
|
| 36 |
+
This step is parameterized, which allows you to configure the step
|
| 37 |
+
independently of the step code, before running it in a pipeline.
|
| 38 |
+
In this example, the step can be configured to use different input data.
|
| 39 |
+
See the documentation for more information:
|
| 40 |
+
|
| 41 |
+
https://docs.zenml.io/user-guide/advanced-guide/configure-steps-pipelines
|
| 42 |
+
|
| 43 |
+
Args:
|
| 44 |
+
dataset_inf: The inference dataset.
|
| 45 |
+
|
| 46 |
+
Returns:
|
| 47 |
+
The predictions as pandas series
|
| 48 |
+
"""
|
| 49 |
+
### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###
|
| 50 |
+
model_version = get_step_context().model_version
|
| 51 |
+
|
| 52 |
+
# run prediction from memory
|
| 53 |
+
predictor = model_version.load_artifact("model")
|
| 54 |
+
predictions = predictor.predict(dataset_inf)
|
| 55 |
+
|
| 56 |
+
predictions = pd.Series(predictions, name="predicted")
|
| 57 |
+
### YOUR CODE ENDS HERE ###
|
| 58 |
+
|
| 59 |
+
return predictions
|
steps/inference_preprocessor.py
ADDED
|
@@ -0,0 +1,52 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# Apache Software License 2.0
|
| 2 |
+
#
|
| 3 |
+
# Copyright (c) ZenML GmbH 2023. All rights reserved.
|
| 4 |
+
#
|
| 5 |
+
# Licensed under the Apache License, Version 2.0 (the "License");
|
| 6 |
+
# you may not use this file except in compliance with the License.
|
| 7 |
+
# You may obtain a copy of the License at
|
| 8 |
+
#
|
| 9 |
+
# http://www.apache.org/licenses/LICENSE-2.0
|
| 10 |
+
#
|
| 11 |
+
# Unless required by applicable law or agreed to in writing, software
|
| 12 |
+
# distributed under the License is distributed on an "AS IS" BASIS,
|
| 13 |
+
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
| 14 |
+
# See the License for the specific language governing permissions and
|
| 15 |
+
# limitations under the License.
|
| 16 |
+
#
|
| 17 |
+
|
| 18 |
+
import pandas as pd
|
| 19 |
+
from sklearn.pipeline import Pipeline
|
| 20 |
+
from typing_extensions import Annotated
|
| 21 |
+
from zenml import step
|
| 22 |
+
|
| 23 |
+
|
| 24 |
+
@step
|
| 25 |
+
def inference_preprocessor(
|
| 26 |
+
dataset_inf: pd.DataFrame,
|
| 27 |
+
preprocess_pipeline: Pipeline,
|
| 28 |
+
target: str,
|
| 29 |
+
) -> Annotated[pd.DataFrame, "inference_dataset"]:
|
| 30 |
+
"""Data preprocessor step.
|
| 31 |
+
|
| 32 |
+
This is an example of a data processor step that prepares the data so that
|
| 33 |
+
it is suitable for model inference. It takes in a dataset as an input step
|
| 34 |
+
artifact and performs any necessary preprocessing steps based on pretrained
|
| 35 |
+
preprocessing pipeline.
|
| 36 |
+
|
| 37 |
+
Args:
|
| 38 |
+
dataset_inf: The inference dataset.
|
| 39 |
+
preprocess_pipeline: Pretrained `Pipeline` to process dataset.
|
| 40 |
+
target: Name of target columns in dataset.
|
| 41 |
+
|
| 42 |
+
Returns:
|
| 43 |
+
The processed dataframe: dataset_inf.
|
| 44 |
+
"""
|
| 45 |
+
### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###
|
| 46 |
+
# artificially adding `target` column to avoid Pipeline issues
|
| 47 |
+
dataset_inf[target] = pd.Series([1] * dataset_inf.shape[0])
|
| 48 |
+
dataset_inf = preprocess_pipeline.transform(dataset_inf)
|
| 49 |
+
dataset_inf.drop(columns=["target"], inplace=True)
|
| 50 |
+
### YOUR CODE ENDS HERE ###
|
| 51 |
+
|
| 52 |
+
return dataset_inf
|
steps/model_evaluator.py
ADDED
|
@@ -0,0 +1,102 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# {% include 'template/license_header' %}
|
| 2 |
+
|
| 3 |
+
import pandas as pd
|
| 4 |
+
import mlflow
|
| 5 |
+
from sklearn.base import ClassifierMixin
|
| 6 |
+
from zenml import step, log_artifact_metadata
|
| 7 |
+
from zenml.client import Client
|
| 8 |
+
from zenml.logger import get_logger
|
| 9 |
+
from zenml import get_step_context
|
| 10 |
+
|
| 11 |
+
logger = get_logger(__name__)
|
| 12 |
+
|
| 13 |
+
experiment_tracker = Client().active_stack.experiment_tracker
|
| 14 |
+
|
| 15 |
+
@step(enable_cache=False, experiment_tracker="mlflow")
|
| 16 |
+
def model_evaluator(
|
| 17 |
+
model: ClassifierMixin,
|
| 18 |
+
dataset_trn: pd.DataFrame,
|
| 19 |
+
dataset_tst: pd.DataFrame,
|
| 20 |
+
min_train_accuracy: float = 0.0,
|
| 21 |
+
min_test_accuracy: float = 0.0,
|
| 22 |
+
) -> float:
|
| 23 |
+
"""Evaluate a trained model.
|
| 24 |
+
|
| 25 |
+
This is an example of a model evaluation step that takes in a model artifact
|
| 26 |
+
previously trained by another step in your pipeline, and a training
|
| 27 |
+
and validation data set pair which it uses to evaluate the model's
|
| 28 |
+
performance. The model metrics are then returned as step output artifacts
|
| 29 |
+
(in this case, the model accuracy on the train and test set).
|
| 30 |
+
|
| 31 |
+
The suggested step implementation also outputs some warnings if the model
|
| 32 |
+
performance does not meet some minimum criteria. This is just an example of
|
| 33 |
+
how you can use steps to monitor your model performance and alert you if
|
| 34 |
+
something goes wrong. As an alternative, you can raise an exception in the
|
| 35 |
+
step to force the pipeline run to fail early and all subsequent steps to
|
| 36 |
+
be skipped.
|
| 37 |
+
|
| 38 |
+
This step is parameterized to configure the step independently of the step code,
|
| 39 |
+
before running it in a pipeline. In this example, the step can be configured
|
| 40 |
+
to use different values for the acceptable model performance thresholds and
|
| 41 |
+
to control whether the pipeline run should fail if the model performance
|
| 42 |
+
does not meet the minimum criteria. See the documentation for more
|
| 43 |
+
information:
|
| 44 |
+
|
| 45 |
+
https://docs.zenml.io/user-guide/advanced-guide/configure-steps-pipelines
|
| 46 |
+
|
| 47 |
+
Args:
|
| 48 |
+
model: The pre-trained model artifact.
|
| 49 |
+
dataset_trn: The train dataset.
|
| 50 |
+
dataset_tst: The test dataset.
|
| 51 |
+
min_train_accuracy: Minimal acceptable training accuracy value.
|
| 52 |
+
min_test_accuracy: Minimal acceptable testing accuracy value.
|
| 53 |
+
fail_on_accuracy_quality_gates: If `True` a `RuntimeException` is raised
|
| 54 |
+
upon not meeting one of the minimal accuracy thresholds.
|
| 55 |
+
|
| 56 |
+
Returns:
|
| 57 |
+
The model accuracy on the test set.
|
| 58 |
+
|
| 59 |
+
Raises:
|
| 60 |
+
RuntimeError: if any of accuracies is lower than respective threshold
|
| 61 |
+
"""
|
| 62 |
+
# context = get_step_context()
|
| 63 |
+
# target = context.inputs["dataset_trn"].run_metadata['target'].value
|
| 64 |
+
target = "target"
|
| 65 |
+
|
| 66 |
+
# Calculate the model accuracy on the train and test set
|
| 67 |
+
trn_acc = model.score(
|
| 68 |
+
dataset_trn.drop(columns=[target]),
|
| 69 |
+
dataset_trn[target],
|
| 70 |
+
)
|
| 71 |
+
logger.info(f"Train accuracy={trn_acc*100:.2f}%")
|
| 72 |
+
tst_acc = model.score(
|
| 73 |
+
dataset_tst.drop(columns=[target]),
|
| 74 |
+
dataset_tst[target],
|
| 75 |
+
)
|
| 76 |
+
logger.info(f"Test accuracy={tst_acc*100:.2f}%")
|
| 77 |
+
|
| 78 |
+
messages = []
|
| 79 |
+
if trn_acc < min_train_accuracy:
|
| 80 |
+
messages.append(
|
| 81 |
+
f"Train accuracy {trn_acc*100:.2f}% is below {min_train_accuracy*100:.2f}% !"
|
| 82 |
+
)
|
| 83 |
+
if tst_acc < min_test_accuracy:
|
| 84 |
+
messages.append(
|
| 85 |
+
f"Test accuracy {tst_acc*100:.2f}% is below {min_test_accuracy*100:.2f}% !"
|
| 86 |
+
)
|
| 87 |
+
else:
|
| 88 |
+
for message in messages:
|
| 89 |
+
logger.warning(message)
|
| 90 |
+
|
| 91 |
+
artifact = get_step_context().model_version.get_artifact("model")
|
| 92 |
+
|
| 93 |
+
log_artifact_metadata(
|
| 94 |
+
metadata={"train_accuracy": float(trn_acc), "test_accuracy": float(tst_acc)},
|
| 95 |
+
artifact_name=artifact.name,
|
| 96 |
+
artifact_version=artifact.version,
|
| 97 |
+
)
|
| 98 |
+
|
| 99 |
+
mlflow.log_metric("train_accuracy", float(trn_acc))
|
| 100 |
+
mlflow.log_metric("test_accuracy", float(tst_acc))
|
| 101 |
+
|
| 102 |
+
return float(trn_acc)
|
steps/model_promoter.py
ADDED
|
@@ -0,0 +1,42 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# {% include 'template/license_header' %}
|
| 2 |
+
|
| 3 |
+
from zenml import get_step_context, step
|
| 4 |
+
from zenml.logger import get_logger
|
| 5 |
+
|
| 6 |
+
logger = get_logger(__name__)
|
| 7 |
+
|
| 8 |
+
|
| 9 |
+
@step
|
| 10 |
+
def model_promoter(accuracy: float, stage: str = "production") -> bool:
|
| 11 |
+
"""Dataset reader step.
|
| 12 |
+
|
| 13 |
+
This is an example of a dataset reader step that load Breast Cancer dataset.
|
| 14 |
+
|
| 15 |
+
This step is parameterized, which allows you to configure the step
|
| 16 |
+
independently of the step code, before running it in a pipeline.
|
| 17 |
+
In this example, the step can be configured with number of rows and logic
|
| 18 |
+
to drop target column or not. See the documentation for more information:
|
| 19 |
+
|
| 20 |
+
https://docs.zenml.io/user-guide/advanced-guide/configure-steps-pipelines
|
| 21 |
+
|
| 22 |
+
Args:
|
| 23 |
+
accuracy: Accuracy of the model.
|
| 24 |
+
stage: Which stage to promote the model to.
|
| 25 |
+
|
| 26 |
+
Returns:
|
| 27 |
+
Whether the model was promoted or not.
|
| 28 |
+
"""
|
| 29 |
+
### ADD YOUR OWN CODE HERE - THIS IS JUST AN EXAMPLE ###
|
| 30 |
+
if accuracy < 0.8:
|
| 31 |
+
logger.info(
|
| 32 |
+
f"Model accuracy {accuracy*100:.2f}% is below 80% ! Not promoting model."
|
| 33 |
+
)
|
| 34 |
+
is_promoted = False
|
| 35 |
+
else:
|
| 36 |
+
logger.info(f"Model promoted to {stage}!")
|
| 37 |
+
is_promoted = True
|
| 38 |
+
model_version = get_step_context().model_version
|
| 39 |
+
model_version.set_stage(stage, force=True)
|
| 40 |
+
|
| 41 |
+
### YOUR CODE ENDS HERE ###
|
| 42 |
+
return is_promoted
|
steps/model_trainer.py
ADDED
|
@@ -0,0 +1,52 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
# {% include 'template/license_header' %}
|
| 2 |
+
|
| 3 |
+
import mlflow
|
| 4 |
+
import pandas as pd
|
| 5 |
+
from sklearn.base import ClassifierMixin
|
| 6 |
+
from sklearn.tree import DecisionTreeClassifier
|
| 7 |
+
from typing_extensions import Annotated
|
| 8 |
+
from zenml import ArtifactConfig, step
|
| 9 |
+
from zenml.client import Client
|
| 10 |
+
from zenml.logger import get_logger
|
| 11 |
+
|
| 12 |
+
logger = get_logger(__name__)
|
| 13 |
+
|
| 14 |
+
experiment_tracker = Client().active_stack.experiment_tracker
|
| 15 |
+
|
| 16 |
+
@step(enable_cache=False, experiment_tracker="mlflow")
|
| 17 |
+
def model_trainer(
|
| 18 |
+
dataset_trn: pd.DataFrame,
|
| 19 |
+
) -> Annotated[ClassifierMixin, ArtifactConfig(name="model", is_model_artifact=True)]:
|
| 20 |
+
"""Configure and train a model on the training dataset.
|
| 21 |
+
|
| 22 |
+
This is an example of a model training step that takes in a dataset artifact
|
| 23 |
+
previously loaded and pre-processed by other steps in your pipeline, then
|
| 24 |
+
configures and trains a model on it. The model is then returned as a step
|
| 25 |
+
output artifact.
|
| 26 |
+
|
| 27 |
+
Args:
|
| 28 |
+
dataset_trn: The preprocessed train dataset.
|
| 29 |
+
target: The name of the target column in the dataset.
|
| 30 |
+
|
| 31 |
+
Returns:
|
| 32 |
+
The trained model artifact.
|
| 33 |
+
"""
|
| 34 |
+
# Use the dataset to fetch the target
|
| 35 |
+
# context = get_step_context()
|
| 36 |
+
# target = context.inputs["dataset_trn"].run_metadata['target'].value
|
| 37 |
+
target = "target"
|
| 38 |
+
|
| 39 |
+
# Initialize the model with the hyperparameters indicated in the step
|
| 40 |
+
# parameters and train it on the training set.
|
| 41 |
+
model = DecisionTreeClassifier()
|
| 42 |
+
logger.info(f"Training model {model}...")
|
| 43 |
+
|
| 44 |
+
model.fit(
|
| 45 |
+
dataset_trn.drop(columns=[target]),
|
| 46 |
+
dataset_trn[target],
|
| 47 |
+
)
|
| 48 |
+
|
| 49 |
+
mlflow.sklearn.log_model(model, "breast_cancer_classifier_model")
|
| 50 |
+
mlflow.sklearn.autolog()
|
| 51 |
+
|
| 52 |
+
return model
|