Spaces:
Runtime error
Runtime error
Upload 10 files
Browse files- .gitattributes +4 -35
- .gitignore +7 -0
- README.md +61 -12
- app.py +771 -0
- dev.py +52 -0
- healthcare_prediction.jpg +0 -0
- requirements.txt +4 -0
- server.py +100 -0
- symptoms_categories.py +197 -0
- utils.py +144 -0
.gitattributes
CHANGED
|
@@ -1,35 +1,4 @@
|
|
| 1 |
-
*.
|
| 2 |
-
*.
|
| 3 |
-
*.
|
| 4 |
-
*.
|
| 5 |
-
*.ckpt filter=lfs diff=lfs merge=lfs -text
|
| 6 |
-
*.ftz filter=lfs diff=lfs merge=lfs -text
|
| 7 |
-
*.gz filter=lfs diff=lfs merge=lfs -text
|
| 8 |
-
*.h5 filter=lfs diff=lfs merge=lfs -text
|
| 9 |
-
*.joblib filter=lfs diff=lfs merge=lfs -text
|
| 10 |
-
*.lfs.* filter=lfs diff=lfs merge=lfs -text
|
| 11 |
-
*.mlmodel filter=lfs diff=lfs merge=lfs -text
|
| 12 |
-
*.model filter=lfs diff=lfs merge=lfs -text
|
| 13 |
-
*.msgpack filter=lfs diff=lfs merge=lfs -text
|
| 14 |
-
*.npy filter=lfs diff=lfs merge=lfs -text
|
| 15 |
-
*.npz filter=lfs diff=lfs merge=lfs -text
|
| 16 |
-
*.onnx filter=lfs diff=lfs merge=lfs -text
|
| 17 |
-
*.ot filter=lfs diff=lfs merge=lfs -text
|
| 18 |
-
*.parquet filter=lfs diff=lfs merge=lfs -text
|
| 19 |
-
*.pb filter=lfs diff=lfs merge=lfs -text
|
| 20 |
-
*.pickle filter=lfs diff=lfs merge=lfs -text
|
| 21 |
-
*.pkl filter=lfs diff=lfs merge=lfs -text
|
| 22 |
-
*.pt filter=lfs diff=lfs merge=lfs -text
|
| 23 |
-
*.pth filter=lfs diff=lfs merge=lfs -text
|
| 24 |
-
*.rar filter=lfs diff=lfs merge=lfs -text
|
| 25 |
-
*.safetensors filter=lfs diff=lfs merge=lfs -text
|
| 26 |
-
saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
| 27 |
-
*.tar.* filter=lfs diff=lfs merge=lfs -text
|
| 28 |
-
*.tar filter=lfs diff=lfs merge=lfs -text
|
| 29 |
-
*.tflite filter=lfs diff=lfs merge=lfs -text
|
| 30 |
-
*.tgz filter=lfs diff=lfs merge=lfs -text
|
| 31 |
-
*.wasm filter=lfs diff=lfs merge=lfs -text
|
| 32 |
-
*.xz filter=lfs diff=lfs merge=lfs -text
|
| 33 |
-
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 34 |
-
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 35 |
-
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
|
|
|
| 1 |
+
*.pkl filter=lfs diff=lfs merge=lfs -text
|
| 2 |
+
*.pt filter=lfs diff=lfs merge=lfs -text
|
| 3 |
+
*.extension filter=lfs diff=lfs merge=lfs -text
|
| 4 |
+
*.bin filter=lfs diff=lfs merge=lfs -text
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
.gitignore
ADDED
|
@@ -0,0 +1,7 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
__pycache__/
|
| 2 |
+
.ipynb_checkpoints
|
| 3 |
+
|
| 4 |
+
.venv
|
| 5 |
+
deployment_files/.*
|
| 6 |
+
deployment_files/client_dir/
|
| 7 |
+
deployment_files/server_dir/
|
README.md
CHANGED
|
@@ -1,12 +1,61 @@
|
|
| 1 |
-
---
|
| 2 |
-
title: Health
|
| 3 |
-
emoji:
|
| 4 |
-
colorFrom:
|
| 5 |
-
colorTo: blue
|
| 6 |
-
sdk: gradio
|
| 7 |
-
sdk_version: 4.44.
|
| 8 |
-
app_file: app.py
|
| 9 |
-
pinned:
|
| 10 |
-
|
| 11 |
-
|
| 12 |
-
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
title: Health Prediction On Encrypted Data Using Fully Homomorphic Encryption
|
| 3 |
+
emoji: 🩺😷
|
| 4 |
+
colorFrom: gray
|
| 5 |
+
colorTo: blue
|
| 6 |
+
sdk: gradio
|
| 7 |
+
sdk_version: 4.44.0
|
| 8 |
+
app_file: app.py
|
| 9 |
+
pinned: true
|
| 10 |
+
tags:
|
| 11 |
+
- FHE
|
| 12 |
+
- PPML
|
| 13 |
+
- privacy
|
| 14 |
+
- privacy preserving machine learning
|
| 15 |
+
- image processing
|
| 16 |
+
- homomorphic encryption
|
| 17 |
+
- security
|
| 18 |
+
python_version: 3.10.6
|
| 19 |
+
---
|
| 20 |
+
|
| 21 |
+
# Healthcare prediction using FHE
|
| 22 |
+
|
| 23 |
+
## Running the application on your machine
|
| 24 |
+
|
| 25 |
+
From this directory, i.e., `health_prediction`, you can proceed with the following steps.
|
| 26 |
+
|
| 27 |
+
### Do once
|
| 28 |
+
|
| 29 |
+
First, create a virtual env and activate it:
|
| 30 |
+
|
| 31 |
+
<!--pytest-codeblocks:skip-->
|
| 32 |
+
|
| 33 |
+
```bash
|
| 34 |
+
python3 -m venv .venv
|
| 35 |
+
source .venv/bin/activate
|
| 36 |
+
```
|
| 37 |
+
|
| 38 |
+
Then, install required packages:
|
| 39 |
+
|
| 40 |
+
<!--pytest-codeblocks:skip-->
|
| 41 |
+
|
| 42 |
+
```bash
|
| 43 |
+
pip3 install pip --upgrade
|
| 44 |
+
pip3 install -U pip wheel setuptools --ignore-installed
|
| 45 |
+
pip3 install -r requirements.txt --ignore-installed
|
| 46 |
+
```
|
| 47 |
+
|
| 48 |
+
## Run the following steps each time you relaunch the application
|
| 49 |
+
|
| 50 |
+
In a terminal, run:
|
| 51 |
+
|
| 52 |
+
<!--pytest-codeblocks:skip-->
|
| 53 |
+
|
| 54 |
+
```bash
|
| 55 |
+
source .venv/bin/activate
|
| 56 |
+
python3 app.py
|
| 57 |
+
```
|
| 58 |
+
|
| 59 |
+
## Interacting with the application
|
| 60 |
+
|
| 61 |
+
Open the given URL link (search for a line like `Running on local URL: http://127.0.0.1:8888/`).
|
app.py
ADDED
|
@@ -0,0 +1,771 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import subprocess
|
| 2 |
+
import time
|
| 3 |
+
from typing import Dict, List, Tuple
|
| 4 |
+
|
| 5 |
+
import gradio as gr # pylint: disable=import-error
|
| 6 |
+
import numpy as np
|
| 7 |
+
import pandas as pd
|
| 8 |
+
import requests
|
| 9 |
+
from symptoms_categories import SYMPTOMS_LIST
|
| 10 |
+
from utils import (
|
| 11 |
+
CLIENT_DIR,
|
| 12 |
+
CURRENT_DIR,
|
| 13 |
+
DEPLOYMENT_DIR,
|
| 14 |
+
INPUT_BROWSER_LIMIT,
|
| 15 |
+
KEYS_DIR,
|
| 16 |
+
SERVER_URL,
|
| 17 |
+
TARGET_COLUMNS,
|
| 18 |
+
TRAINING_FILENAME,
|
| 19 |
+
clean_directory,
|
| 20 |
+
get_disease_name,
|
| 21 |
+
load_data,
|
| 22 |
+
pretty_print,
|
| 23 |
+
)
|
| 24 |
+
|
| 25 |
+
from concrete.ml.deployment import FHEModelClient
|
| 26 |
+
|
| 27 |
+
subprocess.Popen(["uvicorn", "server:app"], cwd=CURRENT_DIR)
|
| 28 |
+
time.sleep(3)
|
| 29 |
+
|
| 30 |
+
# pylint: disable=c-extension-no-member,invalid-name
|
| 31 |
+
|
| 32 |
+
|
| 33 |
+
def is_none(obj) -> bool:
|
| 34 |
+
"""
|
| 35 |
+
Check if the object is None.
|
| 36 |
+
|
| 37 |
+
Args:
|
| 38 |
+
obj (any): The input to be checked.
|
| 39 |
+
|
| 40 |
+
Returns:
|
| 41 |
+
bool: True if the object is None or empty, False otherwise.
|
| 42 |
+
"""
|
| 43 |
+
return obj is None or (obj is not None and len(obj) < 1)
|
| 44 |
+
|
| 45 |
+
|
| 46 |
+
def display_default_symptoms_fn(default_disease: str) -> Dict:
|
| 47 |
+
"""
|
| 48 |
+
Displays the symptoms of a given existing disease.
|
| 49 |
+
|
| 50 |
+
Args:
|
| 51 |
+
default_disease (str): Disease
|
| 52 |
+
Returns:
|
| 53 |
+
Dict: The according symptoms
|
| 54 |
+
"""
|
| 55 |
+
df = pd.read_csv(TRAINING_FILENAME)
|
| 56 |
+
df_filtred = df[df[TARGET_COLUMNS[1]] == default_disease]
|
| 57 |
+
|
| 58 |
+
return {
|
| 59 |
+
default_symptoms: gr.update(
|
| 60 |
+
visible=True,
|
| 61 |
+
value=pretty_print(
|
| 62 |
+
df_filtred.columns[df_filtred.eq(1).any()].to_list(), delimiter=", "
|
| 63 |
+
),
|
| 64 |
+
)
|
| 65 |
+
}
|
| 66 |
+
|
| 67 |
+
|
| 68 |
+
def get_user_symptoms_from_checkboxgroup(checkbox_symptoms: List) -> np.array:
|
| 69 |
+
"""
|
| 70 |
+
Convert the user symptoms into a binary vector representation.
|
| 71 |
+
|
| 72 |
+
Args:
|
| 73 |
+
checkbox_symptoms (List): A list of user symptoms.
|
| 74 |
+
|
| 75 |
+
Returns:
|
| 76 |
+
np.array: A binary vector representing the user's symptoms.
|
| 77 |
+
|
| 78 |
+
Raises:
|
| 79 |
+
KeyError: If a provided symptom is not recognized as a valid symptom.
|
| 80 |
+
|
| 81 |
+
"""
|
| 82 |
+
symptoms_vector = {key: 0 for key in valid_symptoms}
|
| 83 |
+
for pretty_symptom in checkbox_symptoms:
|
| 84 |
+
original_symptom = "_".join((pretty_symptom.lower().split(" ")))
|
| 85 |
+
if original_symptom not in symptoms_vector.keys():
|
| 86 |
+
raise KeyError(
|
| 87 |
+
f"The symptom '{original_symptom}' you provided is not recognized as a valid "
|
| 88 |
+
f"symptom.\nHere is the list of valid symptoms: {symptoms_vector}"
|
| 89 |
+
)
|
| 90 |
+
symptoms_vector[original_symptom] = 1
|
| 91 |
+
|
| 92 |
+
user_symptoms_vect = np.fromiter(symptoms_vector.values(), dtype=float)[np.newaxis, :]
|
| 93 |
+
|
| 94 |
+
assert all(value == 0 or value == 1 for value in user_symptoms_vect.flatten())
|
| 95 |
+
|
| 96 |
+
return user_symptoms_vect
|
| 97 |
+
|
| 98 |
+
|
| 99 |
+
def get_features_fn(*checked_symptoms: Tuple[str]) -> Dict:
|
| 100 |
+
"""
|
| 101 |
+
Get vector features based on the selected symptoms.
|
| 102 |
+
|
| 103 |
+
Args:
|
| 104 |
+
checked_symptoms (Tuple[str]): User symptoms
|
| 105 |
+
|
| 106 |
+
Returns:
|
| 107 |
+
Dict: The encoded user vector symptoms.
|
| 108 |
+
"""
|
| 109 |
+
if not any(lst for lst in checked_symptoms if lst):
|
| 110 |
+
return {
|
| 111 |
+
error_box1: gr.update(visible=True, value="⚠️ Please provide your chief complaints."),
|
| 112 |
+
}
|
| 113 |
+
|
| 114 |
+
if len(pretty_print(checked_symptoms)) < 5:
|
| 115 |
+
print("Provide at least 5 symptoms.")
|
| 116 |
+
return {
|
| 117 |
+
error_box1: gr.update(visible=True, value="⚠️ Provide at least 5 symptoms"),
|
| 118 |
+
one_hot_vect: None,
|
| 119 |
+
}
|
| 120 |
+
|
| 121 |
+
return {
|
| 122 |
+
error_box1: gr.update(visible=False),
|
| 123 |
+
one_hot_vect: gr.update(
|
| 124 |
+
visible=False,
|
| 125 |
+
value=get_user_symptoms_from_checkboxgroup(pretty_print(checked_symptoms)),
|
| 126 |
+
),
|
| 127 |
+
submit_btn: gr.update(value="Data submitted ✅"),
|
| 128 |
+
}
|
| 129 |
+
|
| 130 |
+
|
| 131 |
+
def key_gen_fn(user_symptoms: List[str]) -> Dict:
|
| 132 |
+
"""
|
| 133 |
+
Generate keys for a given user.
|
| 134 |
+
|
| 135 |
+
Args:
|
| 136 |
+
user_symptoms (List[str]): The vector symptoms provided by the user.
|
| 137 |
+
|
| 138 |
+
Returns:
|
| 139 |
+
dict: A dictionary containing the generated keys and related information.
|
| 140 |
+
|
| 141 |
+
"""
|
| 142 |
+
clean_directory()
|
| 143 |
+
|
| 144 |
+
if is_none(user_symptoms):
|
| 145 |
+
print("Error: Please submit your symptoms or select a default disease.")
|
| 146 |
+
return {
|
| 147 |
+
error_box2: gr.update(visible=True, value="⚠️ Please submit your symptoms first."),
|
| 148 |
+
}
|
| 149 |
+
|
| 150 |
+
# Generate a random user ID
|
| 151 |
+
user_id = np.random.randint(0, 2**32)
|
| 152 |
+
print(f"Your user ID is: {user_id}....")
|
| 153 |
+
|
| 154 |
+
client = FHEModelClient(path_dir=DEPLOYMENT_DIR, key_dir=KEYS_DIR / f"{user_id}")
|
| 155 |
+
client.load()
|
| 156 |
+
|
| 157 |
+
# Creates the private and evaluation keys on the client side
|
| 158 |
+
client.generate_private_and_evaluation_keys()
|
| 159 |
+
|
| 160 |
+
# Get the serialized evaluation keys
|
| 161 |
+
serialized_evaluation_keys = client.get_serialized_evaluation_keys()
|
| 162 |
+
assert isinstance(serialized_evaluation_keys, bytes)
|
| 163 |
+
|
| 164 |
+
# Save the evaluation key
|
| 165 |
+
evaluation_key_path = KEYS_DIR / f"{user_id}/evaluation_key"
|
| 166 |
+
with evaluation_key_path.open("wb") as f:
|
| 167 |
+
f.write(serialized_evaluation_keys)
|
| 168 |
+
|
| 169 |
+
serialized_evaluation_keys_shorten_hex = serialized_evaluation_keys.hex()[:INPUT_BROWSER_LIMIT]
|
| 170 |
+
|
| 171 |
+
return {
|
| 172 |
+
error_box2: gr.update(visible=False),
|
| 173 |
+
key_box: gr.update(visible=False, value=serialized_evaluation_keys_shorten_hex),
|
| 174 |
+
user_id_box: gr.update(visible=False, value=user_id),
|
| 175 |
+
key_len_box: gr.update(
|
| 176 |
+
visible=False, value=f"{len(serialized_evaluation_keys) / (10**6):.2f} MB"
|
| 177 |
+
),
|
| 178 |
+
gen_key_btn: gr.update(value="Keys have been generated ✅")
|
| 179 |
+
}
|
| 180 |
+
|
| 181 |
+
|
| 182 |
+
def encrypt_fn(user_symptoms: np.ndarray, user_id: str) -> None:
|
| 183 |
+
"""
|
| 184 |
+
Encrypt the user symptoms vector in the `Client Side`.
|
| 185 |
+
|
| 186 |
+
Args:
|
| 187 |
+
user_symptoms (List[str]): The vector symptoms provided by the user
|
| 188 |
+
user_id (user): The current user's ID
|
| 189 |
+
"""
|
| 190 |
+
|
| 191 |
+
if is_none(user_id) or is_none(user_symptoms):
|
| 192 |
+
print("Error in encryption step: Provide your symptoms and generate the evaluation keys.")
|
| 193 |
+
return {
|
| 194 |
+
error_box3: gr.update(
|
| 195 |
+
visible=True,
|
| 196 |
+
value="⚠️ Please ensure that your symptoms have been submitted and "
|
| 197 |
+
"that you have generated the evaluation key.",
|
| 198 |
+
)
|
| 199 |
+
}
|
| 200 |
+
|
| 201 |
+
# Retrieve the client API
|
| 202 |
+
client = FHEModelClient(path_dir=DEPLOYMENT_DIR, key_dir=KEYS_DIR / f"{user_id}")
|
| 203 |
+
client.load()
|
| 204 |
+
|
| 205 |
+
user_symptoms = np.fromstring(user_symptoms[2:-2], dtype=int, sep=".").reshape(1, -1)
|
| 206 |
+
# quant_user_symptoms = client.model.quantize_input(user_symptoms)
|
| 207 |
+
|
| 208 |
+
encrypted_quantized_user_symptoms = client.quantize_encrypt_serialize(user_symptoms)
|
| 209 |
+
assert isinstance(encrypted_quantized_user_symptoms, bytes)
|
| 210 |
+
encrypted_input_path = KEYS_DIR / f"{user_id}/encrypted_input"
|
| 211 |
+
|
| 212 |
+
with encrypted_input_path.open("wb") as f:
|
| 213 |
+
f.write(encrypted_quantized_user_symptoms)
|
| 214 |
+
|
| 215 |
+
encrypted_quantized_user_symptoms_shorten_hex = encrypted_quantized_user_symptoms.hex()[
|
| 216 |
+
:INPUT_BROWSER_LIMIT
|
| 217 |
+
]
|
| 218 |
+
|
| 219 |
+
return {
|
| 220 |
+
error_box3: gr.update(visible=False),
|
| 221 |
+
one_hot_vect_box: gr.update(visible=True, value=user_symptoms),
|
| 222 |
+
enc_vect_box: gr.update(visible=True, value=encrypted_quantized_user_symptoms_shorten_hex),
|
| 223 |
+
}
|
| 224 |
+
|
| 225 |
+
|
| 226 |
+
def send_input_fn(user_id: str, user_symptoms: np.ndarray) -> Dict:
|
| 227 |
+
"""Send the encrypted data and the evaluation key to the server.
|
| 228 |
+
|
| 229 |
+
Args:
|
| 230 |
+
user_id (str): The current user's ID
|
| 231 |
+
user_symptoms (np.ndarray): The user symptoms
|
| 232 |
+
"""
|
| 233 |
+
|
| 234 |
+
if is_none(user_id) or is_none(user_symptoms):
|
| 235 |
+
return {
|
| 236 |
+
error_box4: gr.update(
|
| 237 |
+
visible=True,
|
| 238 |
+
value="⚠️ Please check your connectivity \n"
|
| 239 |
+
"⚠️ Ensure that the symptoms have been submitted and the evaluation "
|
| 240 |
+
"key has been generated before sending the data to the server.",
|
| 241 |
+
)
|
| 242 |
+
}
|
| 243 |
+
|
| 244 |
+
evaluation_key_path = KEYS_DIR / f"{user_id}/evaluation_key"
|
| 245 |
+
encrypted_input_path = KEYS_DIR / f"{user_id}/encrypted_input"
|
| 246 |
+
|
| 247 |
+
if not evaluation_key_path.is_file():
|
| 248 |
+
print(
|
| 249 |
+
"Error Encountered While Sending Data to the Server: "
|
| 250 |
+
f"The key has been generated correctly - {evaluation_key_path.is_file()=}"
|
| 251 |
+
)
|
| 252 |
+
|
| 253 |
+
return {
|
| 254 |
+
error_box4: gr.update(visible=True, value="⚠️ Please generate the private key first.")
|
| 255 |
+
}
|
| 256 |
+
|
| 257 |
+
if not encrypted_input_path.is_file():
|
| 258 |
+
print(
|
| 259 |
+
"Error Encountered While Sending Data to the Server: The data has not been encrypted "
|
| 260 |
+
f"correctly on the client side - {encrypted_input_path.is_file()=}"
|
| 261 |
+
)
|
| 262 |
+
return {
|
| 263 |
+
error_box4: gr.update(
|
| 264 |
+
visible=True,
|
| 265 |
+
value="⚠️ Please encrypt the data with the private key first.",
|
| 266 |
+
),
|
| 267 |
+
}
|
| 268 |
+
|
| 269 |
+
# Define the data and files to post
|
| 270 |
+
data = {
|
| 271 |
+
"user_id": user_id,
|
| 272 |
+
"input": user_symptoms,
|
| 273 |
+
}
|
| 274 |
+
|
| 275 |
+
files = [
|
| 276 |
+
("files", open(encrypted_input_path, "rb")),
|
| 277 |
+
("files", open(evaluation_key_path, "rb")),
|
| 278 |
+
]
|
| 279 |
+
|
| 280 |
+
# Send the encrypted input and evaluation key to the server
|
| 281 |
+
url = SERVER_URL + "send_input"
|
| 282 |
+
with requests.post(
|
| 283 |
+
url=url,
|
| 284 |
+
data=data,
|
| 285 |
+
files=files,
|
| 286 |
+
) as response:
|
| 287 |
+
print(f"Sending Data: {response.ok=}")
|
| 288 |
+
return {
|
| 289 |
+
error_box4: gr.update(visible=False),
|
| 290 |
+
srv_resp_send_data_box: "Data sent",
|
| 291 |
+
}
|
| 292 |
+
|
| 293 |
+
|
| 294 |
+
def run_fhe_fn(user_id: str) -> Dict:
|
| 295 |
+
"""Send the encrypted input and the evaluation key to the server.
|
| 296 |
+
|
| 297 |
+
Args:
|
| 298 |
+
user_id (int): The current user's ID.
|
| 299 |
+
"""
|
| 300 |
+
if is_none(user_id):
|
| 301 |
+
return {
|
| 302 |
+
error_box5: gr.update(
|
| 303 |
+
visible=True,
|
| 304 |
+
value="⚠️ Please check your connectivity \n"
|
| 305 |
+
"⚠️ Ensure that the symptoms have been submitted, the evaluation "
|
| 306 |
+
"key has been generated and the server received the data "
|
| 307 |
+
"before processing the data.",
|
| 308 |
+
),
|
| 309 |
+
fhe_execution_time_box: None,
|
| 310 |
+
}
|
| 311 |
+
|
| 312 |
+
data = {
|
| 313 |
+
"user_id": user_id,
|
| 314 |
+
}
|
| 315 |
+
|
| 316 |
+
url = SERVER_URL + "run_fhe"
|
| 317 |
+
|
| 318 |
+
with requests.post(
|
| 319 |
+
url=url,
|
| 320 |
+
data=data,
|
| 321 |
+
) as response:
|
| 322 |
+
if not response.ok:
|
| 323 |
+
return {
|
| 324 |
+
error_box5: gr.update(
|
| 325 |
+
visible=True,
|
| 326 |
+
value=(
|
| 327 |
+
"⚠️ An error occurred on the Server Side. "
|
| 328 |
+
"Please check connectivity and data transmission."
|
| 329 |
+
),
|
| 330 |
+
),
|
| 331 |
+
fhe_execution_time_box: gr.update(visible=False),
|
| 332 |
+
}
|
| 333 |
+
else:
|
| 334 |
+
time.sleep(1)
|
| 335 |
+
print(f"response.ok: {response.ok}, {response.json()} - Computed")
|
| 336 |
+
|
| 337 |
+
return {
|
| 338 |
+
error_box5: gr.update(visible=False),
|
| 339 |
+
fhe_execution_time_box: gr.update(visible=True, value=f"{response.json():.2f} seconds"),
|
| 340 |
+
}
|
| 341 |
+
|
| 342 |
+
|
| 343 |
+
def get_output_fn(user_id: str, user_symptoms: np.ndarray) -> Dict:
|
| 344 |
+
"""Retreive the encrypted data from the server.
|
| 345 |
+
|
| 346 |
+
Args:
|
| 347 |
+
user_id (str): The current user's ID
|
| 348 |
+
user_symptoms (np.ndarray): The user symptoms
|
| 349 |
+
"""
|
| 350 |
+
|
| 351 |
+
if is_none(user_id) or is_none(user_symptoms):
|
| 352 |
+
return {
|
| 353 |
+
error_box6: gr.update(
|
| 354 |
+
visible=True,
|
| 355 |
+
value="⚠️ Please check your connectivity \n"
|
| 356 |
+
"⚠️ Ensure that the server has successfully processed and transmitted the data to the client.",
|
| 357 |
+
)
|
| 358 |
+
}
|
| 359 |
+
|
| 360 |
+
data = {
|
| 361 |
+
"user_id": user_id,
|
| 362 |
+
}
|
| 363 |
+
|
| 364 |
+
# Retrieve the encrypted output
|
| 365 |
+
url = SERVER_URL + "get_output"
|
| 366 |
+
with requests.post(
|
| 367 |
+
url=url,
|
| 368 |
+
data=data,
|
| 369 |
+
) as response:
|
| 370 |
+
if response.ok:
|
| 371 |
+
print(f"Receive Data: {response.ok=}")
|
| 372 |
+
|
| 373 |
+
encrypted_output = response.content
|
| 374 |
+
|
| 375 |
+
# Save the encrypted output to bytes in a file as it is too large to pass through
|
| 376 |
+
# regular Gradio buttons (see https://github.com/gradio-app/gradio/issues/1877)
|
| 377 |
+
encrypted_output_path = CLIENT_DIR / f"{user_id}_encrypted_output"
|
| 378 |
+
|
| 379 |
+
with encrypted_output_path.open("wb") as f:
|
| 380 |
+
f.write(encrypted_output)
|
| 381 |
+
return {error_box6: gr.update(visible=False), srv_resp_retrieve_data_box: "Data received"}
|
| 382 |
+
|
| 383 |
+
|
| 384 |
+
def decrypt_fn(
|
| 385 |
+
user_id: str, user_symptoms: np.ndarray, *checked_symptoms, threshold: int = 0.5
|
| 386 |
+
) -> Dict:
|
| 387 |
+
"""Dencrypt the data on the `Client Side`.
|
| 388 |
+
|
| 389 |
+
Args:
|
| 390 |
+
user_id (str): The current user's ID
|
| 391 |
+
user_symptoms (np.ndarray): The user symptoms
|
| 392 |
+
threshold (float): Probability confidence threshold
|
| 393 |
+
|
| 394 |
+
Returns:
|
| 395 |
+
Decrypted output
|
| 396 |
+
"""
|
| 397 |
+
|
| 398 |
+
if is_none(user_id) or is_none(user_symptoms):
|
| 399 |
+
return {
|
| 400 |
+
error_box7: gr.update(
|
| 401 |
+
visible=True,
|
| 402 |
+
value="⚠️ Please check your connectivity \n"
|
| 403 |
+
"⚠️ Ensure that the client has successfully received the data from the server.",
|
| 404 |
+
)
|
| 405 |
+
}
|
| 406 |
+
|
| 407 |
+
# Get the encrypted output path
|
| 408 |
+
encrypted_output_path = CLIENT_DIR / f"{user_id}_encrypted_output"
|
| 409 |
+
|
| 410 |
+
if not encrypted_output_path.is_file():
|
| 411 |
+
print("Error in decryption step: Please run the FHE execution, first.")
|
| 412 |
+
return {
|
| 413 |
+
error_box7: gr.update(
|
| 414 |
+
visible=True,
|
| 415 |
+
value="⚠️ Please ensure that: \n"
|
| 416 |
+
"- the connectivity \n"
|
| 417 |
+
"- the symptoms have been submitted \n"
|
| 418 |
+
"- the evaluation key has been generated \n"
|
| 419 |
+
"- the server processed the encrypted data \n"
|
| 420 |
+
"- the Client received the data from the Server before decrypting the prediction",
|
| 421 |
+
),
|
| 422 |
+
decrypt_box: None,
|
| 423 |
+
}
|
| 424 |
+
|
| 425 |
+
# Load the encrypted output as bytes
|
| 426 |
+
with encrypted_output_path.open("rb") as f:
|
| 427 |
+
encrypted_output = f.read()
|
| 428 |
+
|
| 429 |
+
# Retrieve the client API
|
| 430 |
+
client = FHEModelClient(path_dir=DEPLOYMENT_DIR, key_dir=KEYS_DIR / f"{user_id}")
|
| 431 |
+
client.load()
|
| 432 |
+
|
| 433 |
+
# Deserialize, decrypt and post-process the encrypted output
|
| 434 |
+
output = client.deserialize_decrypt_dequantize(encrypted_output)
|
| 435 |
+
|
| 436 |
+
top3_diseases = np.argsort(output.flatten())[-3:][::-1]
|
| 437 |
+
top3_proba = output[0][top3_diseases]
|
| 438 |
+
|
| 439 |
+
out = ""
|
| 440 |
+
|
| 441 |
+
if top3_proba[0] < threshold or abs(top3_proba[0] - top3_proba[1]) < 0.1:
|
| 442 |
+
out = (
|
| 443 |
+
"⚠️ The prediction appears uncertain; including more symptoms "
|
| 444 |
+
"may improve the results.\n\n"
|
| 445 |
+
)
|
| 446 |
+
|
| 447 |
+
out = (
|
| 448 |
+
f"{out}Given the symptoms you provided: "
|
| 449 |
+
f"{pretty_print(checked_symptoms, case_conversion=str.capitalize, delimiter=', ')}\n\n"
|
| 450 |
+
"Here are the top3 predictions:\n\n"
|
| 451 |
+
f"1. « {get_disease_name(top3_diseases[0])} » with a probability of {top3_proba[0]:.2%}\n"
|
| 452 |
+
f"2. « {get_disease_name(top3_diseases[1])} » with a probability of {top3_proba[1]:.2%}\n"
|
| 453 |
+
f"3. « {get_disease_name(top3_diseases[2])} » with a probability of {top3_proba[2]:.2%}\n"
|
| 454 |
+
)
|
| 455 |
+
|
| 456 |
+
return {
|
| 457 |
+
error_box7: gr.update(visible=False),
|
| 458 |
+
decrypt_box: out,
|
| 459 |
+
submit_btn: gr.update(value="Submit"),
|
| 460 |
+
}
|
| 461 |
+
|
| 462 |
+
|
| 463 |
+
def reset_fn():
|
| 464 |
+
"""Reset the space and clear all the box outputs."""
|
| 465 |
+
|
| 466 |
+
clean_directory()
|
| 467 |
+
|
| 468 |
+
return {
|
| 469 |
+
one_hot_vect: None,
|
| 470 |
+
one_hot_vect_box: None,
|
| 471 |
+
enc_vect_box: gr.update(visible=True, value=None),
|
| 472 |
+
quant_vect_box: gr.update(visible=False, value=None),
|
| 473 |
+
user_id_box: gr.update(visible=False, value=None),
|
| 474 |
+
default_symptoms: gr.update(visible=True, value=None),
|
| 475 |
+
default_disease_box: gr.update(visible=True, value=None),
|
| 476 |
+
key_box: gr.update(visible=True, value=None),
|
| 477 |
+
key_len_box: gr.update(visible=False, value=None),
|
| 478 |
+
fhe_execution_time_box: gr.update(visible=True, value=None),
|
| 479 |
+
decrypt_box: None,
|
| 480 |
+
submit_btn: gr.update(value="Submit"),
|
| 481 |
+
error_box7: gr.update(visible=False),
|
| 482 |
+
error_box1: gr.update(visible=False),
|
| 483 |
+
error_box2: gr.update(visible=False),
|
| 484 |
+
error_box3: gr.update(visible=False),
|
| 485 |
+
error_box4: gr.update(visible=False),
|
| 486 |
+
error_box5: gr.update(visible=False),
|
| 487 |
+
error_box6: gr.update(visible=False),
|
| 488 |
+
srv_resp_send_data_box: None,
|
| 489 |
+
srv_resp_retrieve_data_box: None,
|
| 490 |
+
**{box: None for box in check_boxes},
|
| 491 |
+
}
|
| 492 |
+
|
| 493 |
+
|
| 494 |
+
if __name__ == "__main__":
|
| 495 |
+
|
| 496 |
+
print("Starting demo ...")
|
| 497 |
+
|
| 498 |
+
clean_directory()
|
| 499 |
+
|
| 500 |
+
(X_train, X_test), (y_train, y_test), valid_symptoms, diseases = load_data()
|
| 501 |
+
|
| 502 |
+
with gr.Blocks() as demo:
|
| 503 |
+
|
| 504 |
+
# Link + images
|
| 505 |
+
gr.Markdown()
|
| 506 |
+
gr.Markdown(
|
| 507 |
+
"""
|
| 508 |
+
<p align="center">
|
| 509 |
+
<img width=200 src="https://user-images.githubusercontent.com/5758427/197816413-d9cddad3-ba38-4793-847d-120975e1da11.png">
|
| 510 |
+
</p>
|
| 511 |
+
""")
|
| 512 |
+
gr.Markdown()
|
| 513 |
+
gr.Markdown("""<h2 align="center">Health Prediction On Encrypted Data Using Fully Homomorphic Encryption</h2>""")
|
| 514 |
+
gr.Markdown()
|
| 515 |
+
gr.Markdown(
|
| 516 |
+
"""
|
| 517 |
+
<p align="center">
|
| 518 |
+
<a href="https://github.com/zama-ai/concrete-ml"> <img style="vertical-align: middle; display:inline-block; margin-right: 3px;" width=15 src="https://user-images.githubusercontent.com/5758427/197972109-faaaff3e-10e2-4ab6-80f5-7531f7cfb08f.png">Concrete-ML</a>
|
| 519 |
+
—
|
| 520 |
+
<a href="https://docs.zama.ai/concrete-ml"> <img style="vertical-align: middle; display:inline-block; margin-right: 3px;" width=15 src="https://user-images.githubusercontent.com/5758427/197976802-fddd34c5-f59a-48d0-9bff-7ad1b00cb1fb.png">Documentation</a>
|
| 521 |
+
—
|
| 522 |
+
<a href="https://zama.ai/community"> <img style="vertical-align: middle; display:inline-block; margin-right: 3px;" width=15 src="https://user-images.githubusercontent.com/5758427/197977153-8c9c01a7-451a-4993-8e10-5a6ed5343d02.png">Community</a>
|
| 523 |
+
—
|
| 524 |
+
<a href="https://twitter.com/zama_fhe"> <img style="vertical-align: middle; display:inline-block; margin-right: 3px;" width=15 src="https://user-images.githubusercontent.com/5758427/197975044-bab9d199-e120-433b-b3be-abd73b211a54.png">@zama_fhe</a>
|
| 525 |
+
</p>
|
| 526 |
+
""")
|
| 527 |
+
gr.Markdown()
|
| 528 |
+
gr.Markdown(
|
| 529 |
+
""""
|
| 530 |
+
<p align="center">
|
| 531 |
+
<img width="65%" height="25%" src="https://raw.githubusercontent.com/kcelia/Img/main/healthcare_prediction.jpg">
|
| 532 |
+
</p>
|
| 533 |
+
"""
|
| 534 |
+
)
|
| 535 |
+
gr.Markdown("## Notes")
|
| 536 |
+
gr.Markdown(
|
| 537 |
+
"""
|
| 538 |
+
- The private key is used to encrypt and decrypt the data and shall never be shared.
|
| 539 |
+
- The evaluation key is a public key that the server needs to process encrypted data.
|
| 540 |
+
"""
|
| 541 |
+
)
|
| 542 |
+
|
| 543 |
+
# ------------------------- Step 1 -------------------------
|
| 544 |
+
gr.Markdown("\n")
|
| 545 |
+
gr.Markdown("## Step 1: Select chief complaints")
|
| 546 |
+
gr.Markdown("<hr />")
|
| 547 |
+
gr.Markdown("<span style='color:grey'>Client Side</span>")
|
| 548 |
+
gr.Markdown("Select at least 5 chief complaints from the list below.")
|
| 549 |
+
|
| 550 |
+
# Step 1.1: Provide symptoms
|
| 551 |
+
check_boxes = []
|
| 552 |
+
with gr.Row():
|
| 553 |
+
with gr.Column():
|
| 554 |
+
for category in SYMPTOMS_LIST[:3]:
|
| 555 |
+
with gr.Accordion(pretty_print(category.keys()), open=False):
|
| 556 |
+
check_box = gr.CheckboxGroup(pretty_print(category.values()), show_label=0)
|
| 557 |
+
check_boxes.append(check_box)
|
| 558 |
+
with gr.Column():
|
| 559 |
+
for category in SYMPTOMS_LIST[3:6]:
|
| 560 |
+
with gr.Accordion(pretty_print(category.keys()), open=False):
|
| 561 |
+
check_box = gr.CheckboxGroup(pretty_print(category.values()), show_label=0)
|
| 562 |
+
check_boxes.append(check_box)
|
| 563 |
+
with gr.Column():
|
| 564 |
+
for category in SYMPTOMS_LIST[6:]:
|
| 565 |
+
with gr.Accordion(pretty_print(category.keys()), open=False):
|
| 566 |
+
check_box = gr.CheckboxGroup(pretty_print(category.values()), show_label=0)
|
| 567 |
+
check_boxes.append(check_box)
|
| 568 |
+
|
| 569 |
+
error_box1 = gr.Textbox(label="Error ❌", visible=False)
|
| 570 |
+
|
| 571 |
+
# Default disease, picked from the dataframe
|
| 572 |
+
gr.Markdown(
|
| 573 |
+
"You can choose an **existing disease** and explore its associated symptoms.",
|
| 574 |
+
visible=False,
|
| 575 |
+
)
|
| 576 |
+
|
| 577 |
+
with gr.Row():
|
| 578 |
+
with gr.Column(scale=2):
|
| 579 |
+
default_disease_box = gr.Dropdown(sorted(diseases), label="Diseases", visible=False)
|
| 580 |
+
with gr.Column(scale=5):
|
| 581 |
+
default_symptoms = gr.Textbox(label="Related Symptoms:", visible=False)
|
| 582 |
+
# User vector symptoms encoded in oneHot representation
|
| 583 |
+
one_hot_vect = gr.Textbox(visible=False)
|
| 584 |
+
# Submit botton
|
| 585 |
+
submit_btn = gr.Button("Submit")
|
| 586 |
+
# Clear botton
|
| 587 |
+
clear_button = gr.Button("Reset Space 🔁", visible=False)
|
| 588 |
+
|
| 589 |
+
default_disease_box.change(
|
| 590 |
+
fn=display_default_symptoms_fn, inputs=[default_disease_box], outputs=[default_symptoms]
|
| 591 |
+
)
|
| 592 |
+
|
| 593 |
+
submit_btn.click(
|
| 594 |
+
fn=get_features_fn,
|
| 595 |
+
inputs=[*check_boxes],
|
| 596 |
+
outputs=[one_hot_vect, error_box1, submit_btn],
|
| 597 |
+
)
|
| 598 |
+
|
| 599 |
+
# ------------------------- Step 2 -------------------------
|
| 600 |
+
gr.Markdown("\n")
|
| 601 |
+
gr.Markdown("## Step 2: Encrypt data")
|
| 602 |
+
gr.Markdown("<hr />")
|
| 603 |
+
gr.Markdown("<span style='color:grey'>Client Side</span>")
|
| 604 |
+
# Step 2.1: Key generation
|
| 605 |
+
gr.Markdown(
|
| 606 |
+
"### Key Generation\n\n"
|
| 607 |
+
"In FHE schemes, a secret (enc/dec)ryption keys are generated for encrypting and decrypting data owned by the client. \n\n"
|
| 608 |
+
"Additionally, a public evaluation key is generated, enabling external entities to perform homomorphic operations on encrypted data, without the need to decrypt them. \n\n"
|
| 609 |
+
"The evaluation key will be transmitted to the server for further processing."
|
| 610 |
+
)
|
| 611 |
+
|
| 612 |
+
gen_key_btn = gr.Button("Generate the private and evaluation keys.")
|
| 613 |
+
error_box2 = gr.Textbox(label="Error ❌", visible=False)
|
| 614 |
+
user_id_box = gr.Textbox(label="User ID:", visible=False)
|
| 615 |
+
key_len_box = gr.Textbox(label="Evaluation Key Size:", visible=False)
|
| 616 |
+
key_box = gr.Textbox(label="Evaluation key (truncated):", max_lines=3, visible=False)
|
| 617 |
+
|
| 618 |
+
gen_key_btn.click(
|
| 619 |
+
key_gen_fn,
|
| 620 |
+
inputs=one_hot_vect,
|
| 621 |
+
outputs=[
|
| 622 |
+
key_box,
|
| 623 |
+
user_id_box,
|
| 624 |
+
key_len_box,
|
| 625 |
+
error_box2,
|
| 626 |
+
gen_key_btn,
|
| 627 |
+
],
|
| 628 |
+
)
|
| 629 |
+
|
| 630 |
+
# Step 2.2: Encrypt data locally
|
| 631 |
+
gr.Markdown("### Encrypt the data")
|
| 632 |
+
encrypt_btn = gr.Button("Encrypt the data using the private secret key")
|
| 633 |
+
error_box3 = gr.Textbox(label="Error ❌", visible=False)
|
| 634 |
+
quant_vect_box = gr.Textbox(label="Quantized Vector:", visible=False)
|
| 635 |
+
|
| 636 |
+
with gr.Row():
|
| 637 |
+
with gr.Column():
|
| 638 |
+
one_hot_vect_box = gr.Textbox(label="User Symptoms Vector:", max_lines=10)
|
| 639 |
+
with gr.Column():
|
| 640 |
+
enc_vect_box = gr.Textbox(label="Encrypted Vector:", max_lines=10)
|
| 641 |
+
|
| 642 |
+
encrypt_btn.click(
|
| 643 |
+
encrypt_fn,
|
| 644 |
+
inputs=[one_hot_vect, user_id_box],
|
| 645 |
+
outputs=[
|
| 646 |
+
one_hot_vect_box,
|
| 647 |
+
enc_vect_box,
|
| 648 |
+
error_box3,
|
| 649 |
+
],
|
| 650 |
+
)
|
| 651 |
+
# Step 2.3: Send encrypted data to the server
|
| 652 |
+
gr.Markdown(
|
| 653 |
+
"### Send the encrypted data to the <span style='color:grey'>Server Side</span>"
|
| 654 |
+
)
|
| 655 |
+
error_box4 = gr.Textbox(label="Error ❌", visible=False)
|
| 656 |
+
|
| 657 |
+
# with gr.Row().style(equal_height=False):
|
| 658 |
+
with gr.Row():
|
| 659 |
+
with gr.Column(scale=4):
|
| 660 |
+
send_input_btn = gr.Button("Send data")
|
| 661 |
+
with gr.Column(scale=1):
|
| 662 |
+
srv_resp_send_data_box = gr.Checkbox(label="Data Sent", show_label=False)
|
| 663 |
+
|
| 664 |
+
send_input_btn.click(
|
| 665 |
+
send_input_fn,
|
| 666 |
+
inputs=[user_id_box, one_hot_vect],
|
| 667 |
+
outputs=[error_box4, srv_resp_send_data_box],
|
| 668 |
+
)
|
| 669 |
+
|
| 670 |
+
# ------------------------- Step 3 -------------------------
|
| 671 |
+
gr.Markdown("\n")
|
| 672 |
+
gr.Markdown("## Step 3: Run the FHE evaluation")
|
| 673 |
+
gr.Markdown("<hr />")
|
| 674 |
+
gr.Markdown("<span style='color:grey'>Server Side</span>")
|
| 675 |
+
gr.Markdown(
|
| 676 |
+
"Once the server receives the encrypted data, it can process and compute the output without ever decrypting the data just as it would on clear data.\n\n"
|
| 677 |
+
"This server employs a [Logistic Regression](https://github.com/zama-ai/concrete-ml/tree/release/1.1.x/use_case_examples/disease_prediction) model that has been trained on this [data-set](https://github.com/anujdutt9/Disease-Prediction-from-Symptoms/tree/master/dataset)."
|
| 678 |
+
)
|
| 679 |
+
|
| 680 |
+
run_fhe_btn = gr.Button("Run the FHE evaluation")
|
| 681 |
+
error_box5 = gr.Textbox(label="Error ❌", visible=False)
|
| 682 |
+
fhe_execution_time_box = gr.Textbox(label="Total FHE Execution Time:", visible=True)
|
| 683 |
+
run_fhe_btn.click(
|
| 684 |
+
run_fhe_fn,
|
| 685 |
+
inputs=[user_id_box],
|
| 686 |
+
outputs=[fhe_execution_time_box, error_box5],
|
| 687 |
+
)
|
| 688 |
+
|
| 689 |
+
# ------------------------- Step 4 -------------------------
|
| 690 |
+
gr.Markdown("\n")
|
| 691 |
+
gr.Markdown("## Step 4: Decrypt the data")
|
| 692 |
+
gr.Markdown("<hr />")
|
| 693 |
+
gr.Markdown("<span style='color:grey'>Client Side</span>")
|
| 694 |
+
gr.Markdown(
|
| 695 |
+
"### Get the encrypted data from the <span style='color:grey'>Server Side</span>"
|
| 696 |
+
)
|
| 697 |
+
|
| 698 |
+
error_box6 = gr.Textbox(label="Error ❌", visible=False)
|
| 699 |
+
|
| 700 |
+
# Step 4.1: Data transmission
|
| 701 |
+
# with gr.Row().style(equal_height=True):
|
| 702 |
+
with gr.Row():
|
| 703 |
+
with gr.Column(scale=4):
|
| 704 |
+
get_output_btn = gr.Button("Get data")
|
| 705 |
+
with gr.Column(scale=1):
|
| 706 |
+
srv_resp_retrieve_data_box = gr.Checkbox(label="Data Received", show_label=False)
|
| 707 |
+
|
| 708 |
+
get_output_btn.click(
|
| 709 |
+
get_output_fn,
|
| 710 |
+
inputs=[user_id_box, one_hot_vect],
|
| 711 |
+
outputs=[srv_resp_retrieve_data_box, error_box6],
|
| 712 |
+
)
|
| 713 |
+
|
| 714 |
+
# Step 4.1: Data transmission
|
| 715 |
+
gr.Markdown("### Decrypt the output")
|
| 716 |
+
decrypt_btn = gr.Button("Decrypt the output using the private secret key")
|
| 717 |
+
error_box7 = gr.Textbox(label="Error ❌", visible=False)
|
| 718 |
+
decrypt_box = gr.Textbox(label="Decrypted Output:")
|
| 719 |
+
|
| 720 |
+
decrypt_btn.click(
|
| 721 |
+
decrypt_fn,
|
| 722 |
+
inputs=[user_id_box, one_hot_vect, *check_boxes],
|
| 723 |
+
outputs=[decrypt_box, error_box7, submit_btn],
|
| 724 |
+
)
|
| 725 |
+
|
| 726 |
+
# ------------------------- End -------------------------
|
| 727 |
+
|
| 728 |
+
gr.Markdown(
|
| 729 |
+
"""The app was built with [Concrete ML](https://github.com/zama-ai/concrete-ml), a Privacy-Preserving Machine Learning (PPML) open-source set of tools by Zama.
|
| 730 |
+
Try it yourself and don't forget to star on [Github](https://github.com/zama-ai/concrete-ml) ⭐.
|
| 731 |
+
"""
|
| 732 |
+
)
|
| 733 |
+
|
| 734 |
+
gr.Markdown("\n\n")
|
| 735 |
+
|
| 736 |
+
gr.Markdown(
|
| 737 |
+
"""**Please Note**: This space is intended solely for educational and demonstration purposes.
|
| 738 |
+
It should not be considered as a replacement for professional medical counsel, diagnosis, or therapy for any health or related issues.
|
| 739 |
+
Any questions or concerns about your individual health should be addressed to your doctor or another qualified healthcare provider.
|
| 740 |
+
"""
|
| 741 |
+
)
|
| 742 |
+
|
| 743 |
+
clear_button.click(
|
| 744 |
+
reset_fn,
|
| 745 |
+
outputs=[
|
| 746 |
+
one_hot_vect_box,
|
| 747 |
+
one_hot_vect,
|
| 748 |
+
submit_btn,
|
| 749 |
+
error_box1,
|
| 750 |
+
error_box2,
|
| 751 |
+
error_box3,
|
| 752 |
+
error_box4,
|
| 753 |
+
error_box5,
|
| 754 |
+
error_box6,
|
| 755 |
+
error_box7,
|
| 756 |
+
default_disease_box,
|
| 757 |
+
default_symptoms,
|
| 758 |
+
user_id_box,
|
| 759 |
+
key_len_box,
|
| 760 |
+
key_box,
|
| 761 |
+
quant_vect_box,
|
| 762 |
+
enc_vect_box,
|
| 763 |
+
srv_resp_send_data_box,
|
| 764 |
+
srv_resp_retrieve_data_box,
|
| 765 |
+
fhe_execution_time_box,
|
| 766 |
+
decrypt_box,
|
| 767 |
+
*check_boxes,
|
| 768 |
+
],
|
| 769 |
+
)
|
| 770 |
+
|
| 771 |
+
demo.launch()
|
dev.py
ADDED
|
@@ -0,0 +1,52 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
"""Generating deployment files."""
|
| 2 |
+
|
| 3 |
+
import shutil
|
| 4 |
+
|
| 5 |
+
from pathlib import Path
|
| 6 |
+
|
| 7 |
+
import pandas as pd
|
| 8 |
+
|
| 9 |
+
from concrete.ml.sklearn import LogisticRegression as ConcreteLogisticRegression
|
| 10 |
+
from concrete.ml.deployment import FHEModelDev
|
| 11 |
+
|
| 12 |
+
|
| 13 |
+
# Data files location
|
| 14 |
+
TRAINING_FILE_NAME = "./data/Training_preprocessed.csv"
|
| 15 |
+
TESTING_FILE_NAME = "./data/Testing_preprocessed.csv"
|
| 16 |
+
|
| 17 |
+
# Load data
|
| 18 |
+
df_train = pd.read_csv(TRAINING_FILE_NAME)
|
| 19 |
+
df_test = pd.read_csv(TESTING_FILE_NAME)
|
| 20 |
+
|
| 21 |
+
# Split the data into X_train, y_train, X_test_, y_test sets
|
| 22 |
+
TARGET_COLUMN = ["prognosis_encoded", "prognosis"]
|
| 23 |
+
|
| 24 |
+
y_train = df_train[TARGET_COLUMN[0]].values.flatten()
|
| 25 |
+
y_test = df_test[TARGET_COLUMN[0]].values.flatten()
|
| 26 |
+
|
| 27 |
+
X_train = df_train.drop(TARGET_COLUMN, axis=1)
|
| 28 |
+
X_test = df_test.drop(TARGET_COLUMN, axis=1)
|
| 29 |
+
|
| 30 |
+
# Concrete ML model
|
| 31 |
+
|
| 32 |
+
# Models parameters
|
| 33 |
+
optimal_param = {"C": 0.9, "n_bits": 13, "solver": "sag", "multi_class": "auto"}
|
| 34 |
+
|
| 35 |
+
clf = ConcreteLogisticRegression(**optimal_param)
|
| 36 |
+
|
| 37 |
+
# Fit the model
|
| 38 |
+
clf.fit(X_train, y_train)
|
| 39 |
+
|
| 40 |
+
# Compile the model
|
| 41 |
+
fhe_circuit = clf.compile(X_train)
|
| 42 |
+
|
| 43 |
+
fhe_circuit.client.keygen(force=False)
|
| 44 |
+
|
| 45 |
+
path_to_model = Path("./deployment_files/").resolve()
|
| 46 |
+
|
| 47 |
+
if path_to_model.exists():
|
| 48 |
+
shutil.rmtree(path_to_model)
|
| 49 |
+
|
| 50 |
+
dev = FHEModelDev(path_to_model, clf)
|
| 51 |
+
|
| 52 |
+
dev.save(via_mlir=True)
|
healthcare_prediction.jpg
ADDED
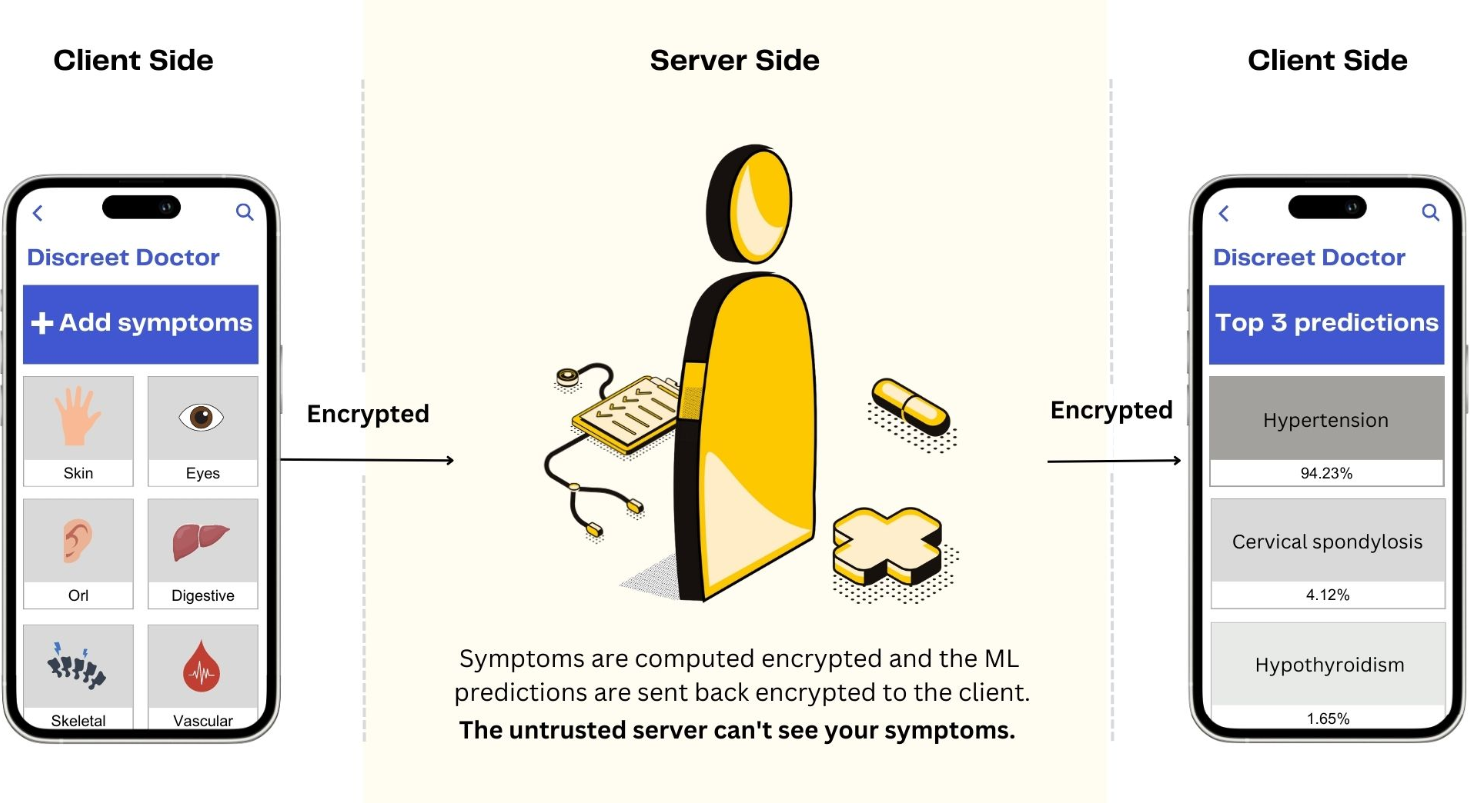
|
requirements.txt
ADDED
|
@@ -0,0 +1,4 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
concrete-ml==1.4.0
|
| 2 |
+
gradio
|
| 3 |
+
uvicorn>=0.21.0
|
| 4 |
+
fastapi>=0.93.0
|
server.py
ADDED
|
@@ -0,0 +1,100 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
"""Server that will listen for GET and POST requests from the client."""
|
| 2 |
+
|
| 3 |
+
import time
|
| 4 |
+
from typing import List
|
| 5 |
+
|
| 6 |
+
from fastapi import FastAPI, File, Form, UploadFile
|
| 7 |
+
from fastapi.responses import JSONResponse, Response
|
| 8 |
+
from utils import DEPLOYMENT_DIR, SERVER_DIR # pylint: disable=no-name-in-module
|
| 9 |
+
|
| 10 |
+
from concrete.ml.deployment import FHEModelServer
|
| 11 |
+
|
| 12 |
+
# Load the FHE server
|
| 13 |
+
FHE_SERVER = FHEModelServer(DEPLOYMENT_DIR)
|
| 14 |
+
|
| 15 |
+
# Initialize an instance of FastAPI
|
| 16 |
+
app = FastAPI()
|
| 17 |
+
|
| 18 |
+
# Define the default route
|
| 19 |
+
@app.get("/")
|
| 20 |
+
def root():
|
| 21 |
+
"""
|
| 22 |
+
Root endpoint of the health prediction API.
|
| 23 |
+
|
| 24 |
+
Returns:
|
| 25 |
+
dict: The welcome message.
|
| 26 |
+
"""
|
| 27 |
+
return {"message": "Welcome to your disease prediction with FHE!"}
|
| 28 |
+
|
| 29 |
+
|
| 30 |
+
@app.post("/send_input")
|
| 31 |
+
def send_input(
|
| 32 |
+
user_id: str = Form(),
|
| 33 |
+
files: List[UploadFile] = File(),
|
| 34 |
+
):
|
| 35 |
+
"""Send the inputs to the server."""
|
| 36 |
+
|
| 37 |
+
print("\nSend the data to the server ............\n")
|
| 38 |
+
|
| 39 |
+
# Receive the Client's files (Evaluation key + Encrypted symptoms)
|
| 40 |
+
evaluation_key_path = SERVER_DIR / f"{user_id}_valuation_key"
|
| 41 |
+
encrypted_input_path = SERVER_DIR / f"{user_id}_encrypted_input"
|
| 42 |
+
|
| 43 |
+
# Save the files using the above paths
|
| 44 |
+
with encrypted_input_path.open("wb") as encrypted_input, evaluation_key_path.open(
|
| 45 |
+
"wb"
|
| 46 |
+
) as evaluation_key:
|
| 47 |
+
encrypted_input.write(files[0].file.read())
|
| 48 |
+
evaluation_key.write(files[1].file.read())
|
| 49 |
+
|
| 50 |
+
|
| 51 |
+
@app.post("/run_fhe")
|
| 52 |
+
def run_fhe(
|
| 53 |
+
user_id: str = Form(),
|
| 54 |
+
):
|
| 55 |
+
"""Inference in FHE."""
|
| 56 |
+
|
| 57 |
+
print("\nRun in FHE in the server ............\n")
|
| 58 |
+
evaluation_key_path = SERVER_DIR / f"{user_id}_valuation_key"
|
| 59 |
+
encrypted_input_path = SERVER_DIR / f"{user_id}_encrypted_input"
|
| 60 |
+
|
| 61 |
+
# Read the files (Evaluation key + Encrypted symptoms) using the above paths
|
| 62 |
+
with encrypted_input_path.open("rb") as encrypted_output_file, evaluation_key_path.open(
|
| 63 |
+
"rb"
|
| 64 |
+
) as evaluation_key_file:
|
| 65 |
+
encrypted_output = encrypted_output_file.read()
|
| 66 |
+
evaluation_key = evaluation_key_file.read()
|
| 67 |
+
|
| 68 |
+
# Run the FHE execution
|
| 69 |
+
start = time.time()
|
| 70 |
+
encrypted_output = FHE_SERVER.run(encrypted_output, evaluation_key)
|
| 71 |
+
assert isinstance(encrypted_output, bytes)
|
| 72 |
+
fhe_execution_time = round(time.time() - start, 2)
|
| 73 |
+
|
| 74 |
+
# Retrieve the encrypted output path
|
| 75 |
+
encrypted_output_path = SERVER_DIR / f"{user_id}_encrypted_output"
|
| 76 |
+
|
| 77 |
+
# Write the file using the above path
|
| 78 |
+
with encrypted_output_path.open("wb") as f:
|
| 79 |
+
f.write(encrypted_output)
|
| 80 |
+
|
| 81 |
+
return JSONResponse(content=fhe_execution_time)
|
| 82 |
+
|
| 83 |
+
|
| 84 |
+
@app.post("/get_output")
|
| 85 |
+
def get_output(user_id: str = Form()):
|
| 86 |
+
"""Retrieve the encrypted output from the server."""
|
| 87 |
+
|
| 88 |
+
print("\nGet the output from the server ............\n")
|
| 89 |
+
|
| 90 |
+
# Path where the encrypted output is saved
|
| 91 |
+
encrypted_output_path = SERVER_DIR / f"{user_id}_encrypted_output"
|
| 92 |
+
|
| 93 |
+
# Read the file using the above path
|
| 94 |
+
with encrypted_output_path.open("rb") as f:
|
| 95 |
+
encrypted_output = f.read()
|
| 96 |
+
|
| 97 |
+
time.sleep(1)
|
| 98 |
+
|
| 99 |
+
# Send the encrypted output
|
| 100 |
+
return Response(encrypted_output)
|
symptoms_categories.py
ADDED
|
@@ -0,0 +1,197 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
"""
|
| 2 |
+
In this file, we roughly split up a list of symptoms, taken from "./training.csv" file, avalaible
|
| 3 |
+
through: "https://github.com/anujdutt9/Disease-Prediction-from-Symptoms/tree/master/dataset"
|
| 4 |
+
into medical categories, in order to make the UI more plesant for the users.
|
| 5 |
+
|
| 6 |
+
Each variable contains a list of symptoms sthat can be pecific to a part of the body or to a list
|
| 7 |
+
of similar symptoms.
|
| 8 |
+
"""
|
| 9 |
+
|
| 10 |
+
|
| 11 |
+
DIGESTIVE_SYSTEM_SYMPTOMS = {
|
| 12 |
+
"DIGESTIVE_SYSTEM_CONCERNS": [
|
| 13 |
+
"stomach_pain",
|
| 14 |
+
"acidity",
|
| 15 |
+
"vomiting",
|
| 16 |
+
"indigestion",
|
| 17 |
+
"constipation",
|
| 18 |
+
"abdominal_pain",
|
| 19 |
+
"diarrhea",
|
| 20 |
+
"nausea",
|
| 21 |
+
"distention_of_abdomen",
|
| 22 |
+
"stomach_bleeding",
|
| 23 |
+
"pain_during_bowel_movements",
|
| 24 |
+
"passage_of_gases",
|
| 25 |
+
"red_spots_over_body",
|
| 26 |
+
"swelling_of_stomach",
|
| 27 |
+
"bloody_stool",
|
| 28 |
+
"irritation_in_anus",
|
| 29 |
+
"pain_in_anal_region",
|
| 30 |
+
"abnormal_menstruation",
|
| 31 |
+
]
|
| 32 |
+
}
|
| 33 |
+
|
| 34 |
+
DERMATOLOGICAL_SYMPTOMS = {
|
| 35 |
+
"DERMATOLOGICAL_CONCERNS": [
|
| 36 |
+
"itching",
|
| 37 |
+
"skin_rash",
|
| 38 |
+
"pus_filled_pimples",
|
| 39 |
+
"blackheads",
|
| 40 |
+
"scurving",
|
| 41 |
+
"skin_peeling",
|
| 42 |
+
"silver_like_dusting",
|
| 43 |
+
"small_dents_in_nails",
|
| 44 |
+
"inflammatory_nails",
|
| 45 |
+
"blister",
|
| 46 |
+
"red_sore_around_nose",
|
| 47 |
+
"bruising",
|
| 48 |
+
"yellow_crust_ooze",
|
| 49 |
+
"dischromic_patches",
|
| 50 |
+
"nodal_skin_eruptions",
|
| 51 |
+
"toxic_look_(typhus)",
|
| 52 |
+
"brittle_nails",
|
| 53 |
+
"yellowish_skin",
|
| 54 |
+
]
|
| 55 |
+
}
|
| 56 |
+
|
| 57 |
+
ORL_SYMPTOMS = {
|
| 58 |
+
"ORL_CONCERNS": [
|
| 59 |
+
"loss_of_smell",
|
| 60 |
+
"continuous_sneezing",
|
| 61 |
+
"runny_nose",
|
| 62 |
+
"patches_in_throat",
|
| 63 |
+
"throat_irritation",
|
| 64 |
+
"sinus_pressure",
|
| 65 |
+
"enlarged_thyroid",
|
| 66 |
+
"loss_of_balance",
|
| 67 |
+
"unsteadiness",
|
| 68 |
+
"dizziness",
|
| 69 |
+
"spinning_movements",
|
| 70 |
+
]
|
| 71 |
+
}
|
| 72 |
+
|
| 73 |
+
THORAX_SYMPTOMS = {
|
| 74 |
+
"THORAX_CONCERNS": [
|
| 75 |
+
"breathlessness",
|
| 76 |
+
"chest_pain",
|
| 77 |
+
"cough",
|
| 78 |
+
"rusty_sputum",
|
| 79 |
+
"phlegm",
|
| 80 |
+
"mucoid_sputum",
|
| 81 |
+
"congestion",
|
| 82 |
+
"blood_in_sputum",
|
| 83 |
+
"fast_heart_rate",
|
| 84 |
+
]
|
| 85 |
+
}
|
| 86 |
+
|
| 87 |
+
OPHTHALMOLOGICAL_SYMPTOMS = {
|
| 88 |
+
"OPHTHALMOLOGICAL_CONCERNS": [
|
| 89 |
+
"sunken_eyes",
|
| 90 |
+
"redness_of_eyes",
|
| 91 |
+
"watering_from_eyes",
|
| 92 |
+
"blurred_and_distorted_vision",
|
| 93 |
+
"pain_behind_the_eyes",
|
| 94 |
+
"visual_disturbances",
|
| 95 |
+
]
|
| 96 |
+
}
|
| 97 |
+
|
| 98 |
+
VASCULAR_LYMPHATIC_SYMPTOMS = {
|
| 99 |
+
"VASCULAR_AND_LYMPHATIC_CONCERNS": [
|
| 100 |
+
"cold_hands_and_feets",
|
| 101 |
+
"swollen_blood_vessels",
|
| 102 |
+
"swollen_legs",
|
| 103 |
+
"swelled_lymph_nodes",
|
| 104 |
+
"palpitations",
|
| 105 |
+
"prominent_veins_on_calf",
|
| 106 |
+
"yellowing_of_eyes",
|
| 107 |
+
"puffy_face_and_eyes",
|
| 108 |
+
"severe_fluid_overload",
|
| 109 |
+
"swollen_extremeties",
|
| 110 |
+
]
|
| 111 |
+
}
|
| 112 |
+
|
| 113 |
+
UROLOGICAL_SYMPTOMS = {
|
| 114 |
+
"UROLOGICAL_CONCERNS": [
|
| 115 |
+
"burning_micturition",
|
| 116 |
+
"spotting_urination",
|
| 117 |
+
"yellow_urine",
|
| 118 |
+
"bladder_discomfort",
|
| 119 |
+
"foul_smell_of_urine",
|
| 120 |
+
"continuous_feel_of_urine",
|
| 121 |
+
"polyuria",
|
| 122 |
+
"dark_urine",
|
| 123 |
+
]
|
| 124 |
+
}
|
| 125 |
+
|
| 126 |
+
MUSCULOSKELETAL_SYMPTOMS = {
|
| 127 |
+
"MUSCULOSKELETAL_CONCERNS": [
|
| 128 |
+
"joint_pain",
|
| 129 |
+
"muscle_wasting",
|
| 130 |
+
"muscle_pain",
|
| 131 |
+
"muscle_weakness",
|
| 132 |
+
"knee_pain",
|
| 133 |
+
"stiff_neck",
|
| 134 |
+
"swelling_joints",
|
| 135 |
+
"movement_stiffness",
|
| 136 |
+
"hip_joint_pain",
|
| 137 |
+
"painful_walking",
|
| 138 |
+
"weakness_of_one_body_side",
|
| 139 |
+
"neck_pain",
|
| 140 |
+
"back_pain",
|
| 141 |
+
"weakness_in_limbs",
|
| 142 |
+
"cramps",
|
| 143 |
+
]
|
| 144 |
+
}
|
| 145 |
+
|
| 146 |
+
GENERAL_SYMPTOMS = {
|
| 147 |
+
"GENERAL_CONCERNS": [
|
| 148 |
+
"acute_liver_failure",
|
| 149 |
+
"anxiety",
|
| 150 |
+
"restlessness",
|
| 151 |
+
"lethargy",
|
| 152 |
+
"mood_swings",
|
| 153 |
+
"irritability",
|
| 154 |
+
"lack_of_concentration",
|
| 155 |
+
"fatigue",
|
| 156 |
+
"malaise",
|
| 157 |
+
"weight_gain",
|
| 158 |
+
"increased_appetite",
|
| 159 |
+
"weight_loss",
|
| 160 |
+
"loss_of_appetite",
|
| 161 |
+
"excess_body_fat",
|
| 162 |
+
"excessive_hunger",
|
| 163 |
+
"ulcers_on_tongue",
|
| 164 |
+
"shivering",
|
| 165 |
+
"chills",
|
| 166 |
+
"irregular_sugar_level",
|
| 167 |
+
"high_fever",
|
| 168 |
+
"slurred_speech",
|
| 169 |
+
"sweating",
|
| 170 |
+
"internal_itching",
|
| 171 |
+
"mild_fever",
|
| 172 |
+
"dehydration",
|
| 173 |
+
"headache",
|
| 174 |
+
"frequent_unprotected_sexual_intercourse_with_multiple_partners",
|
| 175 |
+
"drying_and_tingling_lips",
|
| 176 |
+
"altered_sensorium",
|
| 177 |
+
"family_history",
|
| 178 |
+
"receiving_blood_transfusion",
|
| 179 |
+
"receiving_unsterile_injections",
|
| 180 |
+
"chronic_alcohol_abuse",
|
| 181 |
+
]
|
| 182 |
+
}
|
| 183 |
+
|
| 184 |
+
SYMPTOMS_LIST = [
|
| 185 |
+
# Column 1
|
| 186 |
+
DIGESTIVE_SYSTEM_SYMPTOMS,
|
| 187 |
+
UROLOGICAL_SYMPTOMS,
|
| 188 |
+
VASCULAR_LYMPHATIC_SYMPTOMS,
|
| 189 |
+
# Column 2
|
| 190 |
+
ORL_SYMPTOMS,
|
| 191 |
+
DERMATOLOGICAL_SYMPTOMS,
|
| 192 |
+
MUSCULOSKELETAL_SYMPTOMS,
|
| 193 |
+
# Column 3
|
| 194 |
+
OPHTHALMOLOGICAL_SYMPTOMS,
|
| 195 |
+
THORAX_SYMPTOMS,
|
| 196 |
+
GENERAL_SYMPTOMS,
|
| 197 |
+
]
|
utils.py
ADDED
|
@@ -0,0 +1,144 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
import os
|
| 2 |
+
import shutil
|
| 3 |
+
from pathlib import Path
|
| 4 |
+
from typing import List, Tuple, Union
|
| 5 |
+
|
| 6 |
+
import numpy
|
| 7 |
+
import pandas
|
| 8 |
+
|
| 9 |
+
from concrete.ml.sklearn import XGBClassifier as ConcreteXGBoostClassifier
|
| 10 |
+
|
| 11 |
+
# Max Input to be displayed on the HuggingFace space brower using Gradio
|
| 12 |
+
# Too large inputs, slow down the server: https://github.com/gradio-app/gradio/issues/1877
|
| 13 |
+
INPUT_BROWSER_LIMIT = 380
|
| 14 |
+
|
| 15 |
+
# Store the server's URL
|
| 16 |
+
SERVER_URL = "http://localhost:8000/"
|
| 17 |
+
|
| 18 |
+
CURRENT_DIR = Path(__file__).parent
|
| 19 |
+
DEPLOYMENT_DIR = CURRENT_DIR / "deployment_files"
|
| 20 |
+
KEYS_DIR = DEPLOYMENT_DIR / ".fhe_keys"
|
| 21 |
+
CLIENT_DIR = DEPLOYMENT_DIR / "client_dir"
|
| 22 |
+
SERVER_DIR = DEPLOYMENT_DIR / "server_dir"
|
| 23 |
+
|
| 24 |
+
ALL_DIRS = [KEYS_DIR, CLIENT_DIR, SERVER_DIR]
|
| 25 |
+
|
| 26 |
+
# Columns that define the target
|
| 27 |
+
TARGET_COLUMNS = ["prognosis_encoded", "prognosis"]
|
| 28 |
+
|
| 29 |
+
TRAINING_FILENAME = "./data/Training_preprocessed.csv"
|
| 30 |
+
TESTING_FILENAME = "./data/Testing_preprocessed.csv"
|
| 31 |
+
|
| 32 |
+
# pylint: disable=invalid-name
|
| 33 |
+
|
| 34 |
+
|
| 35 |
+
def pretty_print(
|
| 36 |
+
inputs, case_conversion=str.title, which_replace: str = "_", to_what: str = " ", delimiter=None
|
| 37 |
+
):
|
| 38 |
+
"""
|
| 39 |
+
Prettify and sort the input as a list of string.
|
| 40 |
+
|
| 41 |
+
Args:
|
| 42 |
+
inputs (Any): The inputs to be prettified.
|
| 43 |
+
|
| 44 |
+
Returns:
|
| 45 |
+
List: The prettified and sorted list of inputs.
|
| 46 |
+
|
| 47 |
+
"""
|
| 48 |
+
# Flatten the list if required
|
| 49 |
+
pretty_list = []
|
| 50 |
+
for item in inputs:
|
| 51 |
+
if isinstance(item, list):
|
| 52 |
+
pretty_list.extend(item)
|
| 53 |
+
else:
|
| 54 |
+
pretty_list.append(item)
|
| 55 |
+
|
| 56 |
+
# Sort
|
| 57 |
+
pretty_list = sorted(list(set(pretty_list)))
|
| 58 |
+
# Replace
|
| 59 |
+
pretty_list = [item.replace(which_replace, to_what) for item in pretty_list]
|
| 60 |
+
pretty_list = [case_conversion(item) for item in pretty_list]
|
| 61 |
+
if delimiter:
|
| 62 |
+
pretty_list = f"{delimiter.join(pretty_list)}."
|
| 63 |
+
|
| 64 |
+
return pretty_list
|
| 65 |
+
|
| 66 |
+
|
| 67 |
+
def clean_directory() -> None:
|
| 68 |
+
"""
|
| 69 |
+
Clear direcgtories
|
| 70 |
+
"""
|
| 71 |
+
print("Cleaning...\n")
|
| 72 |
+
for target_dir in ALL_DIRS:
|
| 73 |
+
if os.path.exists(target_dir) and os.path.isdir(target_dir):
|
| 74 |
+
shutil.rmtree(target_dir)
|
| 75 |
+
target_dir.mkdir(exist_ok=True, parents=True)
|
| 76 |
+
|
| 77 |
+
|
| 78 |
+
def get_disease_name(encoded_prediction: int, file_name: str = TRAINING_FILENAME) -> str:
|
| 79 |
+
"""Return the disease name given its encoded label.
|
| 80 |
+
|
| 81 |
+
Args:
|
| 82 |
+
encoded_prediction (int): The encoded prediction
|
| 83 |
+
file_name (str): The data file path
|
| 84 |
+
|
| 85 |
+
Returns:
|
| 86 |
+
str: The according disease name
|
| 87 |
+
"""
|
| 88 |
+
df = pandas.read_csv(file_name, usecols=TARGET_COLUMNS).drop_duplicates()
|
| 89 |
+
disease_name, _ = df[df[TARGET_COLUMNS[0]] == encoded_prediction].values.flatten()
|
| 90 |
+
return disease_name
|
| 91 |
+
|
| 92 |
+
|
| 93 |
+
def load_data() -> Union[Tuple[pandas.DataFrame, numpy.ndarray], List]:
|
| 94 |
+
"""
|
| 95 |
+
Return the data
|
| 96 |
+
|
| 97 |
+
Args:
|
| 98 |
+
None
|
| 99 |
+
|
| 100 |
+
Return:
|
| 101 |
+
The train, testing set and valid symptoms.
|
| 102 |
+
"""
|
| 103 |
+
# Load data
|
| 104 |
+
df_train = pandas.read_csv(TRAINING_FILENAME)
|
| 105 |
+
df_test = pandas.read_csv(TESTING_FILENAME)
|
| 106 |
+
|
| 107 |
+
# Separate the traget from the training / testing set:
|
| 108 |
+
# TARGET_COLUMNS[0] -> "prognosis_encoded" -> contains the numeric label of the disease
|
| 109 |
+
# TARGET_COLUMNS[1] -> "prognosis" -> contains the name of the disease
|
| 110 |
+
|
| 111 |
+
y_train = df_train[TARGET_COLUMNS[0]]
|
| 112 |
+
X_train = df_train.drop(columns=TARGET_COLUMNS, axis=1, errors="ignore")
|
| 113 |
+
|
| 114 |
+
y_test = df_test[TARGET_COLUMNS[0]]
|
| 115 |
+
X_test = df_test.drop(columns=TARGET_COLUMNS, axis=1, errors="ignore")
|
| 116 |
+
|
| 117 |
+
return (
|
| 118 |
+
(X_train, X_test),
|
| 119 |
+
(y_train, y_test),
|
| 120 |
+
X_train.columns.to_list(),
|
| 121 |
+
df_train[TARGET_COLUMNS[1]].unique().tolist(),
|
| 122 |
+
)
|
| 123 |
+
|
| 124 |
+
|
| 125 |
+
def load_model(X_train: pandas.DataFrame, y_train: numpy.ndarray):
|
| 126 |
+
"""
|
| 127 |
+
Load a pre-trained serialized model
|
| 128 |
+
|
| 129 |
+
Args:
|
| 130 |
+
X_train (pandas.DataFrame): Training set
|
| 131 |
+
y_train (numpy.ndarray): Targets of the training set
|
| 132 |
+
|
| 133 |
+
Return:
|
| 134 |
+
The Concrete ML model and its circuit
|
| 135 |
+
"""
|
| 136 |
+
# Parameters
|
| 137 |
+
concrete_args = {"max_depth": 1, "n_bits": 3, "n_estimators": 3, "n_jobs": -1}
|
| 138 |
+
classifier = ConcreteXGBoostClassifier(**concrete_args)
|
| 139 |
+
# Train the model
|
| 140 |
+
classifier.fit(X_train, y_train)
|
| 141 |
+
# Compile the model
|
| 142 |
+
circuit = classifier.compile(X_train)
|
| 143 |
+
|
| 144 |
+
return classifier, circuit
|