metadata
tags:
- feature-extraction
- image-classification
- timm
- biology
- cancer
- owkin
- histology
library_name: timm
model-index:
- name: owkin_pancancer
results:
- task:
type: image-classification
name: Image Classification
dataset:
name: Camelyon16[Meta]
type: image-classification
metrics:
- type: accuracy
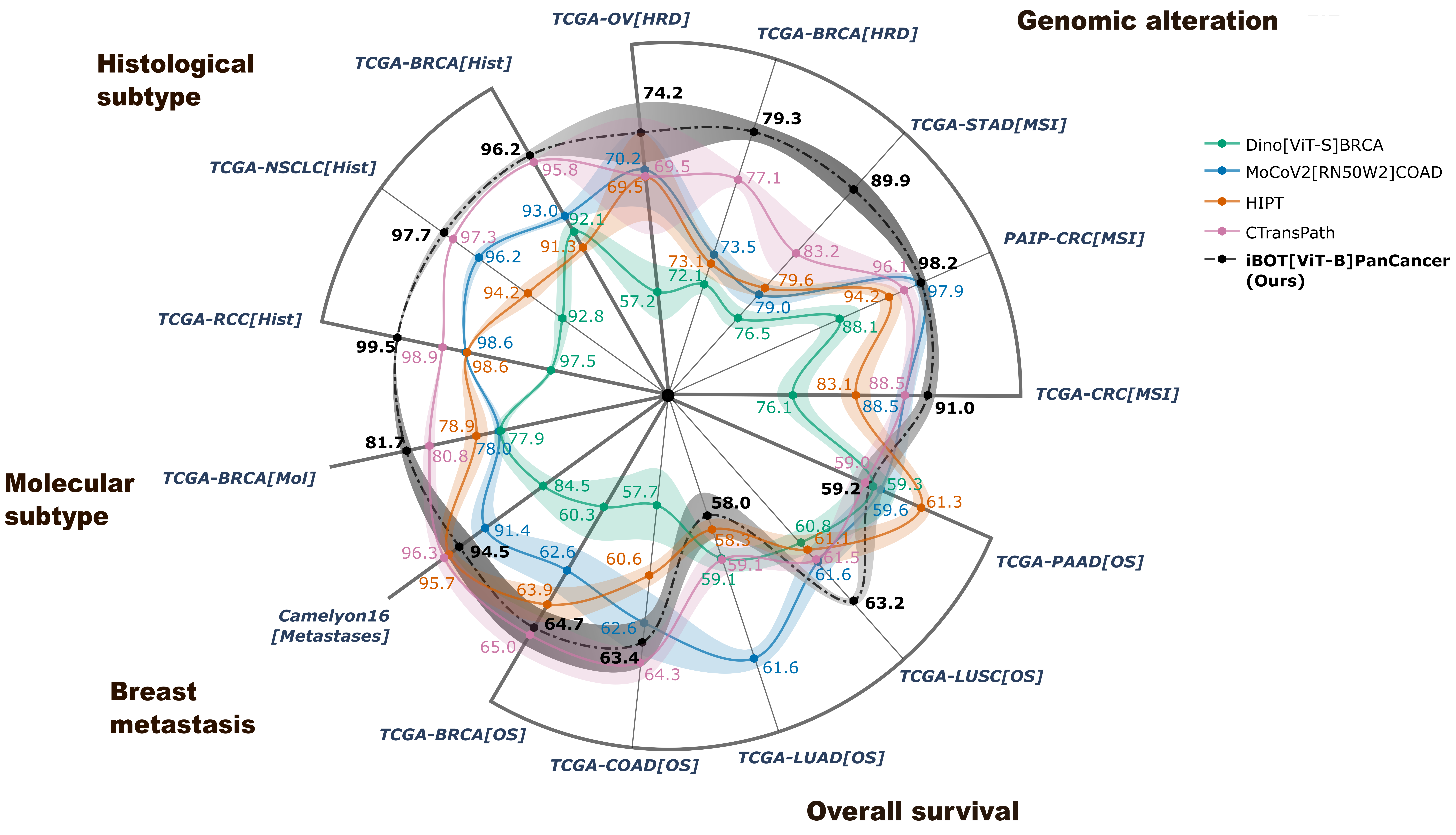
value: 94.5 ± 4.4
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-BRCA[Hist]
type: image-classification
metrics:
- type: accuracy
value: 96.2 ± 3.3
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-BRCA[HRD]
type: image-classification
metrics:
- type: accuracy
value: 79.3 ± 2.4
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-BRCA[Mol]
type: image-classification
metrics:
- type: accuracy
value: 81.7 ± 1.6
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-BRCA[OS]
type: image-classification
metrics:
- type: accuracy
value: 64.7 ± 5.7
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-CRC[MSI]
type: image-classification
metrics:
- type: accuracy
value: 91.0 ± 2.2
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-COAD[OS]
type: image-classification
metrics:
- type: accuracy
value: 63.4 ± 7.4
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-NSCLC[CType]
type: image-classification
metrics:
- type: accuracy
value: 97.7 ± 1.3
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-LUAD[OS]
type: image-classification
metrics:
- type: accuracy
value: 53.8 ± 4.5
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-LUSC[OS]
type: image-classification
metrics:
- type: accuracy
value: 62.2 ± 2.9
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-OV[HRD]
type: image-classification
metrics:
- type: accuracy
value: 74.2 ± 8.6
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-RCC[CType]
type: image-classification
metrics:
- type: accuracy
value: 99.5 ± 0.2
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-STAD[MSI]
type: image-classification
metrics:
- type: accuracy
value: 89.9 ± 3.9
name: ROC AUC
verified: false
- task:
type: image-classification
name: Image Classification
dataset:
name: TCGA-PAAD[OS]
type: image-classification
metrics:
- type: accuracy
value: 59.2 ± 4.1
name: ROC AUC
verified: false
widget:
- src: https://github.com/owkin/HistoSSLscaling/raw/main/assets/example.tif
example_title: pancancer tile
co2_eq_emissions:
emissions: 14590
source: https://www.medrxiv.org/content/10.1101/2023.07.21.23292757v2
training_type: pre-training
geographical_location: Jean Zay cluster, France (~40 gCO₂eq/kWh)
hardware_used: 32 V100 32Gb GPUs, 1216 GPU hours
license: other
pipeline_tag: feature-extraction
inference: false
datasets:
- owkin/camelyon16-features
- owkin/nct-crc-he
- 1aurent/NCT-CRC-HE
metrics:
- roc_auc
Model card for vit_base_patch16_224.owkin_pancancer
A Vision Transformer (ViT) image classification model.
Trained by Owkin on 40M pan-cancer histology tiles from TCGA.
Model Details
- Model Type: Image classification / feature backbone
- Model Stats:
- Params (M): 85.8
- Image size: 224 x 224 x 3
- Papers:
- Scaling Self-Supervised Learning for Histopathology with Masked Image Modeling: https://www.medrxiv.org/content/10.1101/2023.07.21.23292757v2
- Dataset: TGCA: https://portal.gdc.cancer.gov/
- Original: https://github.com/owkin/HistoSSLscaling/
- License: https://github.com/owkin/HistoSSLscaling/blob/main/LICENSE.txt
Model Usage
Image Embeddings
from urllib.request import urlopen
from PIL import Image
import timm
# get example histology image
img = Image.open(
urlopen(
"https://github.com/owkin/HistoSSLscaling/raw/main/assets/example.tif"
)
)
# load model from the hub
model = timm.create_model(
model_name="hf-hub:1aurent/vit_base_patch16_224.owkin_pancancer",
pretrained=True,
).eval()
# get model specific transforms (normalization, resize)
data_config = timm.data.resolve_model_data_config(model)
transforms = timm.data.create_transform(**data_config, is_training=False)
output = model(transforms(img).unsqueeze(0)) # output is (batch_size, num_features) shaped tensor
Citation
@article {Filiot2023.07.21.23292757,
author = {Alexandre Filiot and Ridouane Ghermi and Antoine Olivier and Paul Jacob and Lucas Fidon and Alice Mac Kain and Charlie Saillard and Jean-Baptiste Schiratti},
title = {Scaling Self-Supervised Learning for Histopathology with Masked Image Modeling},
elocation-id = {2023.07.21.23292757},
year = {2023},
doi = {10.1101/2023.07.21.23292757},
publisher = {Cold Spring Harbor Laboratory Press},
URL = {https://www.medrxiv.org/content/early/2023/09/14/2023.07.21.23292757},
eprint = {https://www.medrxiv.org/content/early/2023/09/14/2023.07.21.23292757.full.pdf},
journal = {medRxiv}
}