license: cc-by-4.0
Protein Sequence Modelling with Bayesian Flow Networks
Welcome to the model weights for the paper "Protein Sequence Modelling with Bayesian Flow Networks". Using the code on our GitHub page, you can sample from our trained models ProtBFN, for general proteins, and AbBFN, for antibody VH chains.
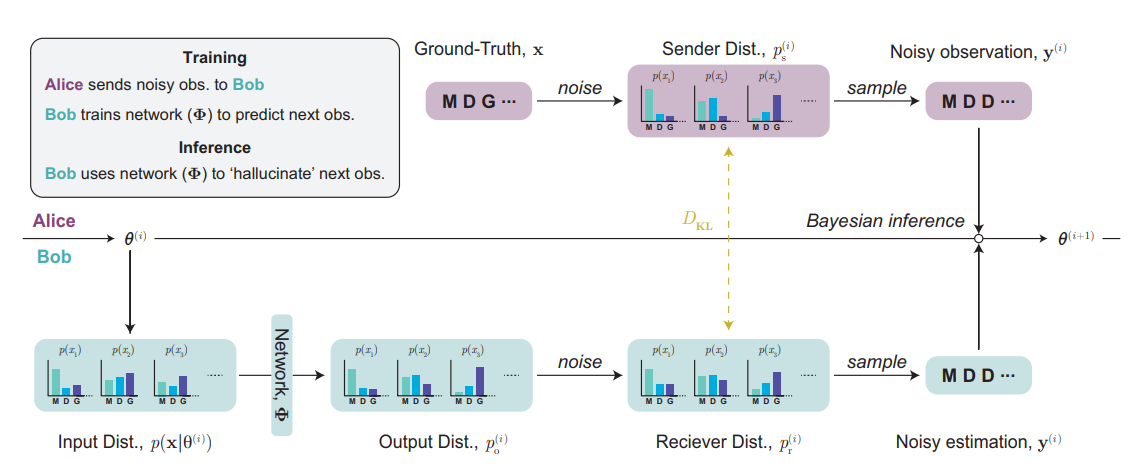
Bayesian Flow Networks are a new approach to generative modelling, and can be viewed as an extension of diffusion models to the parameter space of probability distributions. They define a continuous-time process that maps between a naive prior distribution and a psuedo-deterministic posterior distribution for each variable independently. By training our neural network to 'denoise' the current posterior, by taking into account mutual information between variables, we implicitly minimise a variational lower bound. We can then use our trained neural network to generate samples from the learned distribution.
One of the benefits of defining such a process in probability parameter space is that it can be applied to any family of distributions with continous-valued parameters. This means that BFNs can be directly applied to discrete data, allowing for diffusion-like generative modelling for sequences without restrictive left-to-right inductive biases or relying on discrete-time stochastic processes. The main focus of our work is to investigate the application of BFNs to protein sequences, as represented by a sequence of amino acids. The ProtBFN methodology is broadly summarised below:
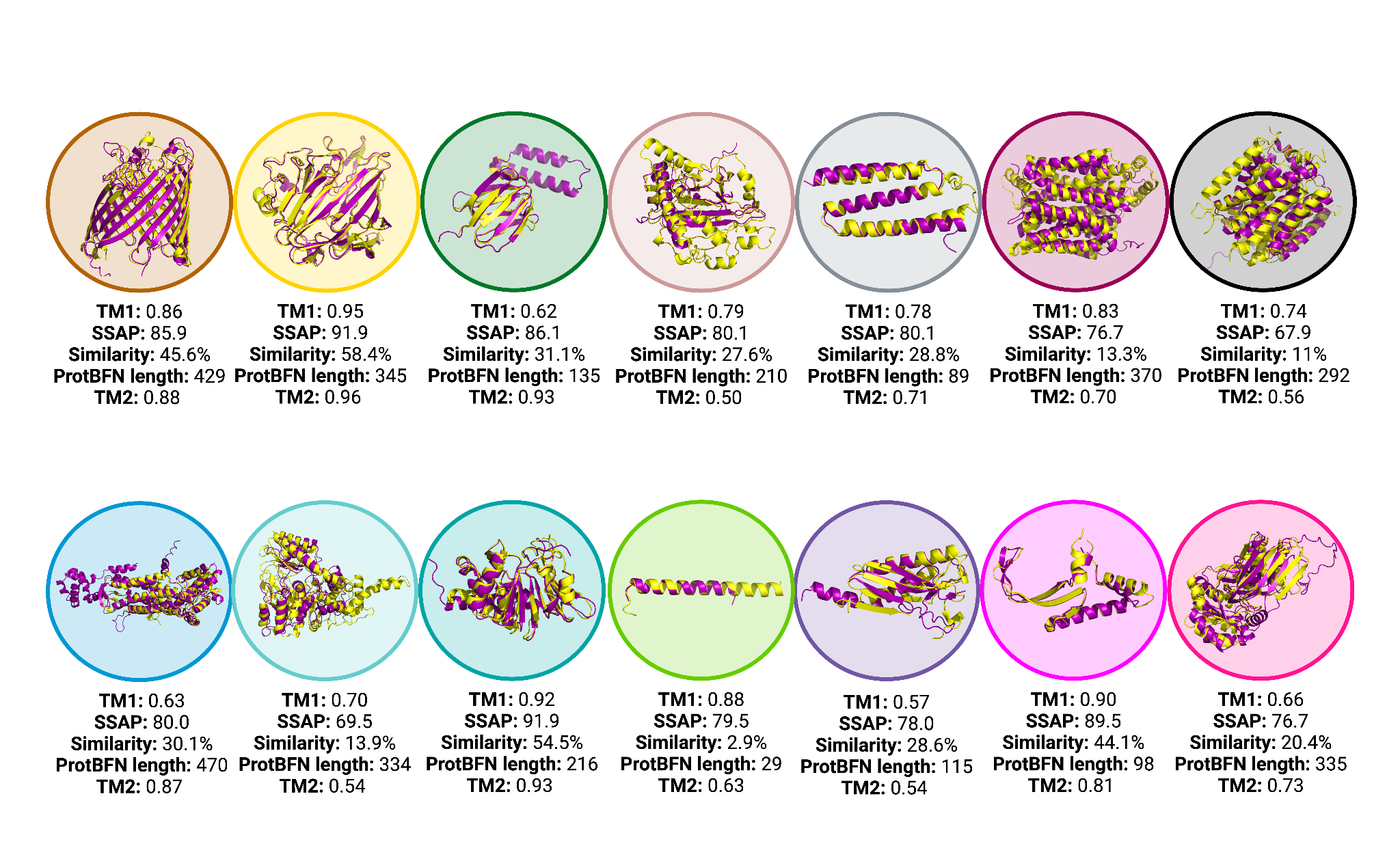
Having trained ProtBFN, we find that it is exceptionally performant at unconditional generation of de novo protein sequences. For example, we find that we are able to rediscover a variety of structural motifs, according to structures predicted by ESMFold, with high sequence novelty:
Cite our work
If you have used ProtBFN or AbBFN in your work, you can cite us using the following bibtex entry:
@article {Atkinson2024.09.24.614734,
author = {Atkinson, Timothy and Barrett, Thomas D. and Cameron, Scott and Guloglu, Bora and Greenig, Matthew and Robinson, Louis and Graves, Alex and Copoiu, Liviu and Laterre, Alexandre},
title = {Protein Sequence Modelling with Bayesian Flow Networks},
elocation-id = {2024.09.24.614734},
year = {2024},
doi = {10.1101/2024.09.24.614734},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2024/09/26/2024.09.24.614734},
eprint = {https://www.biorxiv.org/content/early/2024/09/26/2024.09.24.614734.full.pdf},
journal = {bioRxiv}
}