The dataset viewer is not available for this dataset.
Error code: ConfigNamesError
Exception: BadZipFile
Message: zipfiles that span multiple disks are not supported
Traceback: Traceback (most recent call last):
File "/src/services/worker/src/worker/job_runners/dataset/config_names.py", line 79, in compute_config_names_response
config_names = get_dataset_config_names(
File "/src/services/worker/.venv/lib/python3.9/site-packages/datasets/inspect.py", line 347, in get_dataset_config_names
dataset_module = dataset_module_factory(
File "/src/services/worker/.venv/lib/python3.9/site-packages/datasets/load.py", line 1910, in dataset_module_factory
raise e1 from None
File "/src/services/worker/.venv/lib/python3.9/site-packages/datasets/load.py", line 1885, in dataset_module_factory
return HubDatasetModuleFactoryWithoutScript(
File "/src/services/worker/.venv/lib/python3.9/site-packages/datasets/load.py", line 1270, in get_module
module_name, default_builder_kwargs = infer_module_for_data_files(
File "/src/services/worker/.venv/lib/python3.9/site-packages/datasets/load.py", line 589, in infer_module_for_data_files
split_modules = {
File "/src/services/worker/.venv/lib/python3.9/site-packages/datasets/load.py", line 590, in <dictcomp>
split: infer_module_for_data_files_list(data_files_list, download_config=download_config)
File "/src/services/worker/.venv/lib/python3.9/site-packages/datasets/load.py", line 531, in infer_module_for_data_files_list
return infer_module_for_data_files_list_in_archives(data_files_list, download_config=download_config)
File "/src/services/worker/.venv/lib/python3.9/site-packages/datasets/load.py", line 559, in infer_module_for_data_files_list_in_archives
for f in xglob(extracted, recursive=True, download_config=download_config)[
File "/src/services/worker/.venv/lib/python3.9/site-packages/datasets/utils/file_utils.py", line 1291, in xglob
fs, *_ = url_to_fs(urlpath, **storage_options)
File "/src/services/worker/.venv/lib/python3.9/site-packages/fsspec/core.py", line 395, in url_to_fs
fs = filesystem(protocol, **inkwargs)
File "/src/services/worker/.venv/lib/python3.9/site-packages/fsspec/registry.py", line 293, in filesystem
return cls(**storage_options)
File "/src/services/worker/.venv/lib/python3.9/site-packages/fsspec/spec.py", line 80, in __call__
obj = super().__call__(*args, **kwargs)
File "/src/services/worker/.venv/lib/python3.9/site-packages/fsspec/implementations/zip.py", line 62, in __init__
self.zip = zipfile.ZipFile(
File "/usr/local/lib/python3.9/zipfile.py", line 1266, in __init__
self._RealGetContents()
File "/usr/local/lib/python3.9/zipfile.py", line 1329, in _RealGetContents
endrec = _EndRecData(fp)
File "/usr/local/lib/python3.9/zipfile.py", line 286, in _EndRecData
return _EndRecData64(fpin, -sizeEndCentDir, endrec)
File "/usr/local/lib/python3.9/zipfile.py", line 232, in _EndRecData64
raise BadZipFile("zipfiles that span multiple disks are not supported")
zipfile.BadZipFile: zipfiles that span multiple disks are not supportedNeed help to make the dataset viewer work? Make sure to review how to configure the dataset viewer, and open a discussion for direct support.
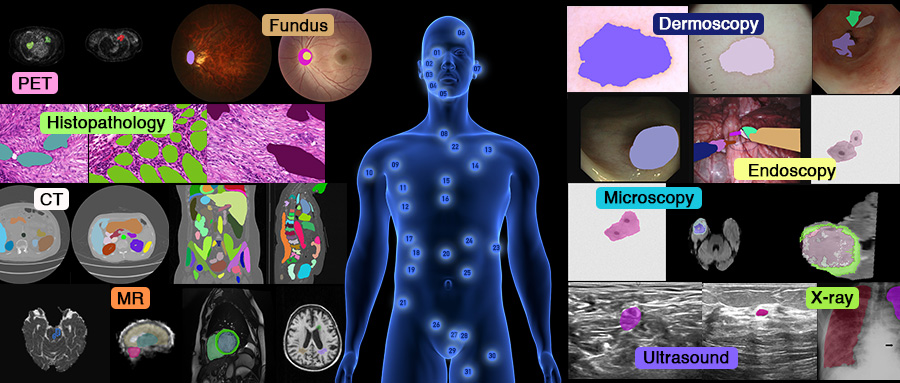
SA-Med2D-20M
The largest benchmark dataset for segmentation in the field of medical imaging.
As is well known, the emergence of ImageNet has greatly propelled the development of AI, especially deep learning. It has provided massive data and powerful baseline models for the computer vision community, enabling researchers to achieve breakthroughs in tasks such as natural image classification, segmentation, and detection. However, in the medical image realm, there lack of such a large dataset for developing powerful medical models.
To address the gap in the medical field, we are introducing the largest benchmark dataset for medical image segmentation. This initiative aims to drive the rapid development of AI in healthcare and accelerate the transformation of computational medicine towards a more inclusive direction.
Please visit the GitHub page and further exploit the dataset!
Due to data privacy and ethical requirements, we currently only provide access to a 16M dataset. We will keep updating and maintaining this database. Please stay tuned for further updates from us.
👉 Filesystem Hierarchy
~/SAM-Med2D-20M
├── images
| ├── mr_00--ACDC--patient001_frame01--x_0006.png
| ├── mr_t1--BraTS2021--BraTS2021_00218--z_0141.png
| ├── ...
| ├── ct_00--CAD_PE--001--x_0125.png
| ├── x_ray--covid_19_ct_cxr--16660_5_1--2d_none.png
|
├── masks
| ├── mr_00--ACDC--patient001_frame01--x_0006--0000_000.png
| ├── mr_t1--BraTS2021--BraTS2021_00218--z_0141--0011_000.png
| ├── ...
| ├── ct_00--CAD_PE--001--x_0125--0000_002.png
| ├── x_ray--covid_19_ct_cxr--16660_5_1--2d_none--0000_001.png
|
├── SAMed2D_v1_class_mapping_id.json
|
├── SAMed2D_v1.json
The SA-Med2D-20M dataset is named following the convention below:
-images
-{modality_sub-modality}--{dataset name}--{ori name}--{dimension_slice}.png
-masks
-{modality_sub-modality}--{dataset name}--{ori name}--{dimension_slice}--{class instance_id}.png
Note: "sub-modality" applies only to 3D data, and when "sub-modality" is "00," it indicates either the absence of a sub-modality or an unknown sub-modality type. "dataset name" refers to the specific dataset name that the case is from. "ori name" is the original case name in its dataset. "dimension slice", e.g., "x_100", indicates the dimension along which we split a 3D case as well as the slice ID in this dimension. If we split a 3D case with axis x and the current slice is 100, then the term can be "x_0100". For 2D datasets, the "dimension_slice id" is uniformly set to "2d_none". "class instance_id", unique to masks, encapsulates both category information and instance id, and the detailed information is stored in the "SAMed2D_v1_class_mapping_id.json" file. For instance, if the category "liver" is assigned the ID "0003" and there is only one instance of this category in the case, the "class instance_id" can be denoted as "0003_000". Besides, the category "liver" in the "SAMed2D_v1_class_mapping_id.json" file is formulated as key-value pair with python-dict format: {"liver": "0003"}.
The file "SAMed2D_v1_class_mapping_id.json" stores the information for converting class instances. The file "SAMed2D_v1.json" contains the path information for all image and mask pairs.
👉 Unzipping split zip files
Windows:
decompress SA-Med2D-16M.zip to automatically extract the other volumes together.
Linux:
1. zip SA-Med2D-16M.zip SA-Med2D-16M.z0* SA-Med2D-16M.z10 -s=0 --out {full}.zip
2. unzip {full}.zip
🤝 免责声明
- SA-Med2D-20M是由多个公开的数据集组成,旨在取之于社区,回馈于社区,为研究人员和开发者提供一个用于学术和技术研究的资源。使用本数据集的任何个人或组织(以下统称为“使用者”)需遵守以下免责声明:
- 数据集来源:本数据集由多个公开的数据集组成,这些数据集的来源已在预印版论文中明确标明。使用者应当遵守原始数据集的相关许可和使用条款。
- 数据准确性:尽管我们已经努力确保数据集的准确性和完整性,但无法对数据集的准确性作出保证。使用者应自行承担使用数据集可能带来的风险和责任。
- 责任限制:在任何情况下,数据集的提供者及相关贡献者均不对使用者的任何行为或结果承担责任。
- 使用约束:使用者在使用本数据集时,应遵守适用的法律法规和伦理规范。使用者不得将本数据集用于非法、侵犯隐私、诽谤、歧视或其他违法或不道德的目的。
- 知识产权:本数据集的知识产权归原始数据集的相关权利人所有,使用者不得以任何方式侵犯数据集的知识产权。
作为非盈利机构,团队倡导和谐友好的开源交流环境,若在开源数据集内发现有侵犯您合法权益的内容,可发送邮件至(yejin@pilab.org.cn, chengjunlong@pilab.org.cn),邮件中请写明侵权相关事实的详细描述并向我们提供相关的权属证明资料。我们将于3个工作日内启动调查处理机制,并采取必要的措施进行处置(如下架相关数据)。但应确保您投诉的真实性,否则采取措施后所产生的不利后果应由您独立承担。
通过下载、复制、访问或使用本数据集,即表示使用者已阅读、理解并同意遵守本免责声明中的所有条款和条件。如果使用者无法接受本免责声明的任何部分,请勿使用本数据集。
🤝 Disclaimer
- SA-Med2D-20M is composed of multiple publicly available datasets and aims to provide a resource for academic and technical research to researchers and developers. Any individual or organization (hereinafter referred to as "User") using this dataset must comply with the following disclaimer:
- Dataset Source: SA-Med2D-20M is composed of multiple publicly available datasets, and the sources of these datasets have been clearly indicated in the preprint paper. Users should adhere to the relevant licenses and terms of use of the original datasets.
- Data Accuracy: While efforts have been made to ensure the accuracy and completeness of the dataset, no guarantee can be given regarding its accuracy. Users assume all risks and liabilities associated with the use of the dataset.
- Limitation of Liability: Under no circumstances shall the dataset providers or contributors be held liable for any actions or outcomes of the Users.
- Usage Constraints: Users must comply with applicable laws, regulations, and ethical norms when using this dataset. The dataset must not be used for illegal, privacy-infringing, defamatory, discriminatory, or other unlawful or unethical purposes.
- Intellectual Property: The intellectual property rights of this dataset belong to the relevant rights holders of the original datasets. Users must not infringe upon the intellectual property rights of the dataset in any way.
As a non-profit organization, we advocate for a harmonious and friendly open-source communication environment. If any content in the open dataset is found to infringe upon your legitimate rights and interests, you can send an email to (yejin@pilab.org.cn, chengjunlong@pilab.org.cn) with a detailed description of the infringement and provide relevant ownership proof materials. We will initiate an investigation and handling mechanism within three working days and take necessary measures (such as removing relevant data) if warranted. However, the authenticity of your complaint must be ensured, as any adverse consequences resulting from the measures taken shall be borne solely by you.
By downloading, copying, accessing, or using this dataset, the User indicates that they have read, understood, and agreed to comply with all the terms and conditions of this disclaimer. If the User cannot accept any part of this disclaimer, please refrain from using this dataset.
🤝 Acknowledgement
- We thank all medical workers and dataset owners for making public datasets available to the community. If you find that your dataset is included in our SA-Med2D-20M but you do not want us to do so, please contact us to remove it.
👋 Hiring & Global Collaboration
- Hiring: We are hiring researchers, engineers, and interns in General Vision Group, Shanghai AI Lab. If you are interested in Medical Foundation Models and General Medical AI, including designing benchmark datasets, general models, evaluation systems, and efficient tools, please contact us.
- Global Collaboration: We're on a mission to redefine medical research, aiming for a more universally adaptable model. Our passionate team is delving into foundational healthcare models, promoting the development of the medical community. Collaborate with us to increase competitiveness, reduce risk, and expand markets.
- Contact: Junjun He(hejunjun@pjlab.org.cn), Jin Ye(yejin@pjlab.org.cn), and Tianbin Li (litianbin@pjlab.org.cn).
👉 Typos of paper
- Formula (1) is incorrect, after correction:
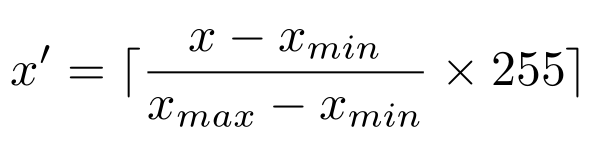
Reference
@misc{ye2023samed2d20m,
title={SA-Med2D-20M Dataset: Segment Anything in 2D Medical Imaging with 20 Million masks},
author={Jin Ye and Junlong Cheng and Jianpin Chen and Zhongying Deng and Tianbin Li and Haoyu Wang and Yanzhou Su and Ziyan Huang and Jilong Chen and Lei Jiang and Hui Sun and Min Zhu and Shaoting Zhang and Junjun He and Yu Qiao},
year={2023},
eprint={2311.11969},
archivePrefix={arXiv},
primaryClass={eess.IV}
}
@misc{cheng2023sammed2d,
title={SAM-Med2D},
author={Junlong Cheng and Jin Ye and Zhongying Deng and Jianpin Chen and Tianbin Li and Haoyu Wang and Yanzhou Su and
Ziyan Huang and Jilong Chen and Lei Jiangand Hui Sun and Junjun He and Shaoting Zhang and Min Zhu and Yu Qiao},
year={2023},
eprint={2308.16184},
archivePrefix={arXiv},
primaryClass={cs.CV}
}
- Downloads last month
- 264