Datasets:
File size: 8,220 Bytes
d36d263 |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 |
---
language: rna
tags:
- Biology
- RNA
license:
- agpl-3.0
size_categories:
- 100K<n<1M
source_datasets:
- multimolecule/crw
- multimolecule/tmrna_website
- multimolecule/srpdb
- multimolecule/spr
- multimolecule/rnp
- multimolecule/rfam
- multimolecule/pdb
task_categories:
- text-generation
- fill-mask
task_ids:
- language-modeling
- masked-language-modeling
pretty_name: bpRNA-1m
library_name: multimolecule
---
# bpRNA-1m
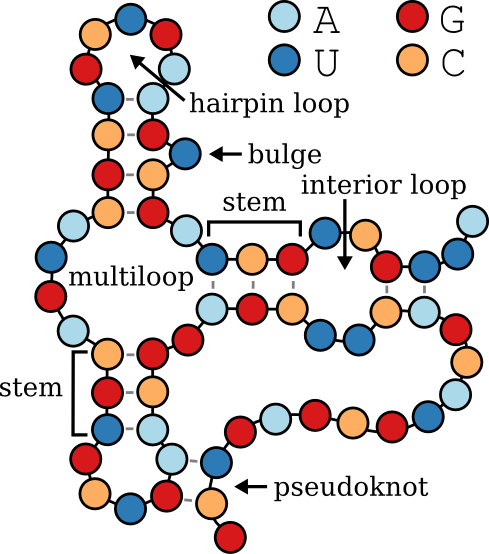
bpRNA-1m is a database of single molecule secondary structures annotated using bpRNA.
## Disclaimer
This is an UNOFFICIAL release of the [bpRNA-1m](https://bprna.cgrb.oregonstate.edu/index.html) by Center for Quantitative Life Sciences of the Oregon State University.
**The team releasing bpRNA did not write this dataset card for this dataset so this dataset card has been written by the MultiMolecule team.**
## Dataset Description
- **Homepage**: https://multimolecule.danling.org/datasets/bprna
- **datasets**: https://huggingface.co/datasets/multimolecule/bprna
- **Point of Contact**: [Center for Quantitative Life Sciences of the Oregon State University](https://cqls.oregonstate.edu)
- **Original URL**: https://bprna.cgrb.oregonstate.edu/index.html
## Example Entry
| id | sequence | secondary_structure | structural_annotation | functional_annotation |
| ------------- | ---------------------------------------------- | --------------------------------------------------- | -------------------------------------------------- | ---------------------------------------------- |
| bpRNA_CRW_170 | GGCUCACCAAGGCGACGACGGGUAGCCGGCCUGAGAGGGCGAC... | ((((((((....))))...))))....((((((..........)))))... | EEEEEEEEEEEEEEEEEEEEEEEESSSSSSSSHHHHSSSSBBBSSSS... | NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN |
## Column Description
The converted dataset consists of the following columns, each providing specific information about the RNA secondary structures, consistent with the bpRNA standard:
- **id**:
A unique identifier for each RNA entry. This ID is derived from the original `.sta` file name and serves as a reference to the specific RNA structure within the dataset.
- **sequence**:
The nucleotide sequence of the RNA molecule, represented using the standard RNA bases:
- **A**: Adenine
- **C**: Cytosine
- **G**: Guanine
- **U**: Uracil
- **secondary_structure**:
The secondary structure of the RNA represented in dot-bracket notation, using up to three types of symbols to indicate base pairing and unpaired regions, as per bpRNA's standard:
- **Dots (`.`)**: Represent unpaired nucleotides.
- **Parentheses (`(` and `)`)**: Represent base pairs in standard stems (page 1).
- **Square Brackets (`[` and `]`)**: Represent base pairs in pseudoknots (page 2).
- **Curly Braces (`{` and `}`)**: Represent base pairs in additional pseudoknots (page 3).
- **structural_annotation**:
Structural annotations categorizing different regions of the RNA based on their roles within the secondary structure, consistent with bpRNA standards:
- **E**: **External Loop** – Regions that are unpaired and external to any loop or helix.
- **S**: **Stem** – Paired regions forming helical structures.
- **H**: **Hairpin Loop** – Unpaired regions at the end of a stem, forming a loop.
- **I**: **Internal Loop** – Unpaired regions between two stems.
- **M**: **Multi-loop** – Junctions where three or more stems converge.
- **B**: **Bulge** – Unpaired nucleotides on one side of a stem.
- **X**: **Ambiguous** or **Undetermined** – Regions where the structure is unknown or cannot be classified.
- **K**: **Pseudoknot** – Regions involved in pseudoknots, where base pairs cross each other.
- **functional_annotation**:
Functional annotations indicating specific functional elements or regions within the RNA sequence, as defined by bpRNA:
- **N**: **None** – No specific functional annotation is assigned.
- **K**: **Pseudoknot** – Marks nucleotides involved in pseudoknot structures, which can be functionally significant.
## Variations
This dataset is available in two variants:
- [bpRNA-1m](https://huggingface.co/datasets/multimolecule/bprna): The main bpRNA-1m dataset.
- [bpRNA-1m(90)](https://huggingface.co/datasets/multimolecule/bprna-90): bpRNA_1m(90) is a subset of bpRNA_1m containing RNAs with less than 90% sequence similarity.
## License
This dataset is licensed under the [AGPL-3.0 License](https://www.gnu.org/licenses/agpl-3.0.html).
```spdx
SPDX-License-Identifier: AGPL-3.0-or-later
```
## Citation
```bibtex
@article{danaee2018bprna,
author = {Danaee, Padideh and Rouches, Mason and Wiley, Michelle and Deng, Dezhong and Huang, Liang and Hendrix, David},
journal = {Nucleic Acids Research},
month = jun,
number = 11,
pages = {5381--5394},
title = {{bpRNA}: large-scale automated annotation and analysis of {RNA} secondary structure},
volume = 46,
year = 2018
}
@article{cannone2002comparative,
author = {Cannone, Jamie J and Subramanian, Sankar and Schnare, Murray N and Collett, James R and D'Souza, Lisa M and Du, Yushi and Feng, Brian and Lin, Nan and Madabusi, Lakshmi V and M{\"u}ller, Kirsten M and Pande, Nupur and Shang, Zhidi and Yu, Nan and Gutell, Robin R},
copyright = {https://www.springernature.com/gp/researchers/text-and-data-mining},
journal = {BMC Bioinformatics},
month = jan,
number = 1,
pages = {2},
publisher = {Springer Science and Business Media LLC},
title = {The comparative {RNA} web ({CRW}) site: an online database of comparative sequence and structure information for ribosomal, intron, and other {RNAs}},
volume = 3,
year = 2002
}
@article{zwieb2003tmrdb,
author = {Zwieb, Christian and Gorodkin, Jan and Knudsen, Bjarne and Burks, Jody and Wower, Jacek},
journal = {Nucleic Acids Research},
month = jan,
number = 1,
pages = {446--447},
publisher = {Oxford University Press (OUP)},
title = {{tmRDB} ({tmRNA} database)},
volume = 31,
year = 2003
}
@article{rosenblad2003srpdb,
author = {Rosenblad, Magnus Alm and Gorodkin, Jan and Knudsen, Bjarne and Zwieb, Christian and Samuelsson, Tore},
journal = {Nucleic Acids Research},
month = jan,
number = 1,
pages = {363--364},
publisher = {Oxford University Press (OUP)},
title = {{SRPDB}: Signal Recognition Particle Database},
volume = 31,
year = 2003
}
@article{sprinzl2005compilation,
author = {Sprinzl, Mathias and Vassilenko, Konstantin S},
journal = {Nucleic Acids Research},
month = jan,
number = {Database issue},
pages = {D139--40},
publisher = {Oxford University Press (OUP)},
title = {Compilation of {tRNA} sequences and sequences of {tRNA} genes},
volume = 33,
year = 2005
}
@article{brown1994ribonuclease,
author = {Brown, J W and Haas, E S and Gilbert, D G and Pace, N R},
journal = {Nucleic Acids Research},
month = sep,
number = 17,
pages = {3660--3662},
publisher = {Oxford University Press (OUP)},
title = {The Ribonuclease {P} database},
volume = 22,
year = 1994
}
@article{griffiths2003rfam,
author = {Griffiths-Jones, Sam and Bateman, Alex and Marshall, Mhairi and Khanna, Ajay and Eddy, Sean R},
journal = {Nucleic Acids Research},
month = jan,
number = 1,
pages = {439--441},
publisher = {Oxford University Press (OUP)},
title = {Rfam: an {RNA} family database},
volume = 31,
year = 2003
}
@article{berman2000protein,
author = {Berman, H M and Westbrook, J and Feng, Z and Gilliland, G and Bhat, T N and Weissig, H and Shindyalov, I N and Bourne, P E},
journal = {Nucleic Acids Research},
month = jan,
number = 1,
pages = {235--242},
publisher = {Oxford University Press (OUP)},
title = {The Protein Data Bank},
volume = 28,
year = 2000
}
```
|