AIDO.Cell-100M
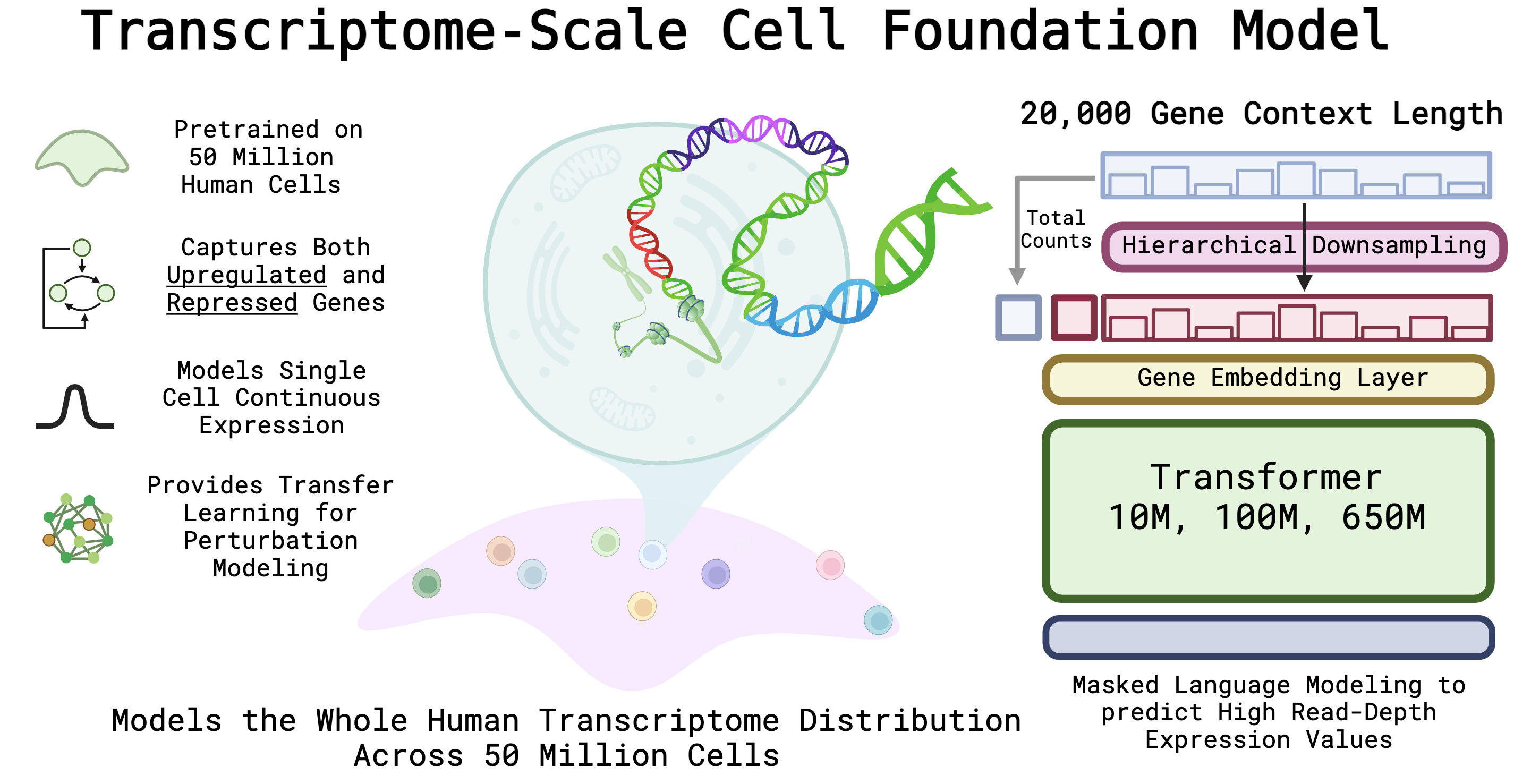
AIDO.Cell-100M is our SOTA cellular foundation model trained on 50 million cells over a diverse set of human tissues and organs. The AIDO.Cell models are capable of handling the entire human transcriptome as input, thus learning accurate and general representations of the human cell's entire transcriptional context. AIDO.Cell achieves state-of-the-art results in tasks such as zero-shot clustering, cell-type classification, and perturbation modeling.
Model Architectural Details
AIDO.Cell uses an auto-discretization strategy for encoding continuous gene expression values, and uses a bidirectional transformer encoder as its backbone. To learn semantically meaningful representations, we employed an BERT-style encoder-only dense transformer architecture. We make minor updates to this architecture to align with current best practices, including using SwiGLU and LayerNorms. Below are more details about the model architecture:
| Model | Layers | Hidden | Heads | Intermediate Hidden Size |
|---|---|---|---|---|
| 3M | 6 | 128 | 4 | 320 |
| 10M | 8 | 256 | 8 | 640 |
| 100M | 18 | 650 | 20 | 1664 |
| 650M | 32 | 1280 | 20 | 3392 |
Pre-training of AIDO.Cell
Here we briefly introduce the details of pre-training of AIDIO.Cell. For more detailed information, please refer to our paper). AIDO.Cell uses the Read Depth-Aware (RDA) pretraining objective where a single cell expression is downsampled into a low read depth, and the model learns to predict the expression count of higher read depth of masked genes.
Data
AIDO.Cell was pretrained on a diverse dataset of 50 million cells from over 100 tissue types. We followed the list of data curated by scFoundation in the supplementary. This list includes datasets from the Gene Expression Omnibus (GEO), the Deeply Integrated human Single-Cell Omnics data (DISCO), the human ensemble cell atlas (hECA), Single Cell Portal and more. After preprocessing and quality control, the training dataset contained 50 million cells, or 963 total billion gene tokens. We partitioned the dataset to set aside 100,000 cells as our validation set.
Training Details
We trained our models with bfloat-16 precision to optimize on memory and speed. The training took place over 256 H100 GPUs over three days for the 100M, and eight days for the 650M version.
Evaluation of AIDO.Cell
We evaluated AIDO.Cell on a series of both zero shots and fine tuned tasks in single cell genomics. For more detailed information, please refer to our paper).
How to Use
For more information, visit: Model Generator.
See experiments/AIDO.Cell
Citation
Please cite AIDO.Cell using the following BibTeX code:
@inproceedings{ho_scaling_2024,
title = {Scaling Dense Representations for Single Cell with Transcriptome-Scale Context},
url = {https://www.biorxiv.org/content/10.1101/2024.11.28.625303v1},
doi = {10.1101/2024.11.28.625303},
publisher = {bioRxiv},
author = {Ho, Nicholas and Ellington, Caleb N. and Hou, Jinyu and Addagudi, Sohan and Mo, Shentong and Tao, Tianhua and Li, Dian and Zhuang, Yonghao and Wang, Hongyi and Cheng, Xingyi and Song, Le and Xing, Eric P.},
year = {2024},
booktitle={NeurIPS 2024 Workshop on AI for New Drug Modalities},
}
- Downloads last month
- 301