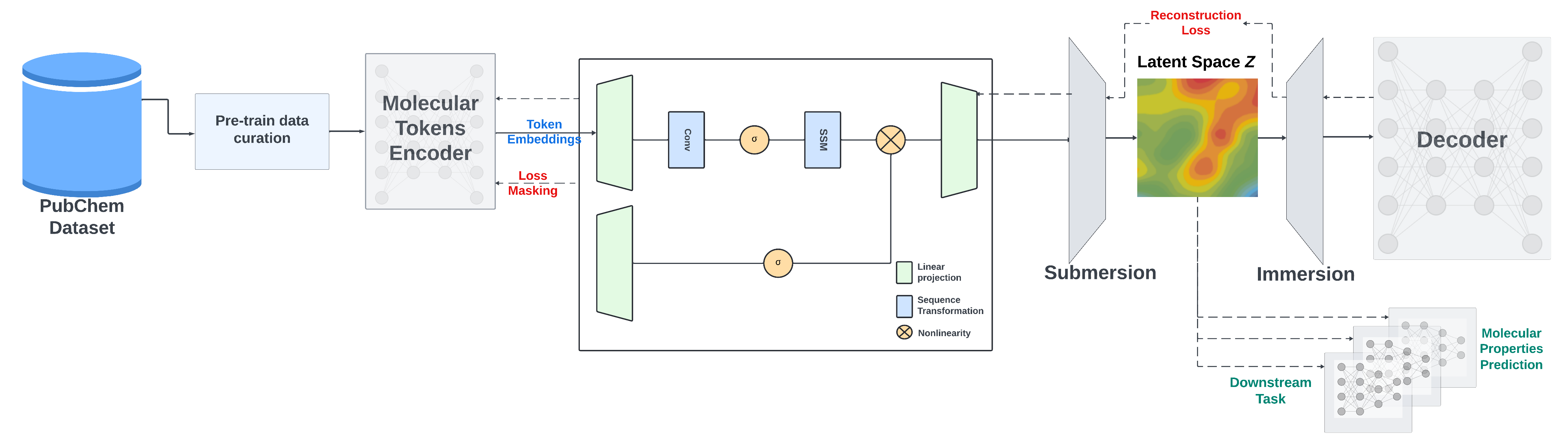
SMILES-based State-Space Encoder-Decoder (SMI-SSED)
This repository provides PyTorch source code associated with our publication, "A Mamba-Based Foundation Model for Chemistry".
Paper: Arxiv Link
HuggingFace: HuggingFace Link
For more information contact: eduardo.soares@ibm.com or evital@br.ibm.com.
Introduction
We present a large encoder-decoder chemical foundation model, SMILES-based State-Space Encoder-Decoder (SMI-SSED), pre-trained on a curated dataset of 91 million SMILES samples sourced from PubChem, equivalent to 4 billion molecular tokens. SMI-SSED supports various complex tasks, including quantum property prediction, with two main variants ($336$ and $8 \times 336M$). Our experiments across multiple benchmark datasets demonstrate state-of-the-art performance for various tasks. Model weights are available at: HuggingFace Link.
Table of Contents
Getting Started
This code and environment have been tested on Nvidia V100s and Nvidia A100s
Pretrained Models and Training Logs
We provide checkpoints of the SMI-SSED model pre-trained on a dataset of ~91M molecules curated from PubChem. The pre-trained model shows competitive performance on classification and regression benchmarks from MoleculeNet. For model weights: HuggingFace Link
Add the SMI-SSED pre-trained weights.pt to the inference/ or finetune/ directory according to your needs. The directory structure should look like the following:
inference/
├── smi_ssed
│ ├── smi_ssed.pt
│ ├── bert_vocab_curated.txt
│ └── load.py
and/or:
finetune/
├── smi_ssed
│ ├── smi_ssed.pt
│ ├── bert_vocab_curated.txt
│ └── load.py
Replicating Conda Environment
Follow these steps to replicate our Conda environment and install the necessary libraries:
Create and Activate Conda Environment
conda create --name smi-ssed-env python=3.10
conda activate smi-ssed-env
Install Packages with Conda
conda install pytorch=2.1.0 pytorch-cuda=11.8 -c pytorch -c nvidia
Install Packages with Pip
pip install -r requirements.txt
Pretraining
For pretraining, we use two strategies: the masked language model method to train the encoder part and an encoder-decoder strategy to refine SMILES reconstruction and improve the generated latent space.
SMI-SSED is pre-trained on canonicalized and curated 91M SMILES from PubChem with the following constraints:
- Compounds are filtered to a maximum length of 202 tokens during preprocessing.
- A 95/5/0 split is used for encoder training, with 5% of the data for decoder pretraining.
- A 100/0/0 split is also used to train the encoder and decoder directly, enhancing model performance.
The pretraining code provides examples of data processing and model training on a smaller dataset, requiring 8 A100 GPUs.
To pre-train the SMI-SSED model, run:
bash training/run_model_training.sh
Use train_model_D.py to train only the decoder or train_model_ED.py to train both the encoder and decoder.
Finetuning
The finetuning datasets and environment can be found in the finetune directory. After setting up the environment, you can run a finetuning task with:
bash finetune/smi_ssed/esol/run_finetune_esol.sh
Finetuning training/checkpointing resources will be available in directories named checkpoint_<measure_name>.
Feature Extraction
The example notebook smi_ssed_encoder_decoder_example.ipynb contains code to load checkpoint files and use the pre-trained model for encoder and decoder tasks. It also includes examples of classification and regression tasks. For model weights: HuggingFace Link
To load smi-ssed, you can simply use:
model = load_smi_ssed(
folder='../inference/smi_ssed',
ckpt_filename='smi_ssed.pt'
)
To encode SMILES into embeddings, you can use:
with torch.no_grad():
encoded_embeddings = model.encode(df['SMILES'], return_torch=True)
For decoder, you can use the function, so you can return from embeddings to SMILES strings:
with torch.no_grad():
decoded_smiles = model.decode(encoded_embeddings)