metadata
license: mit
library_name: sklearn
tags:
- sklearn
- skops
- tabular-classification
widget:
structuredData:
area error:
- 30.29
- 96.05
- 48.31
compactness error:
- 0.01911
- 0.01652
- 0.01484
concave points error:
- 0.01037
- 0.0137
- 0.01093
concavity error:
- 0.02701
- 0.02269
- 0.02813
fractal dimension error:
- 0.003586
- 0.001698
- 0.002461
mean area:
- 481.9
- 1130
- 748.9
mean compactness:
- 0.1058
- 0.1029
- 0.1223
mean concave points:
- 0.03821
- 0.07951
- 0.08087
mean concavity:
- 0.08005
- 0.108
- 0.1466
mean fractal dimension:
- 0.06373
- 0.05461
- 0.05796
mean perimeter:
- 81.09
- 123.6
- 101.7
mean radius:
- 12.47
- 18.94
- 15.46
mean smoothness:
- 0.09965
- 0.09009
- 0.1092
mean symmetry:
- 0.1925
- 0.1582
- 0.1931
mean texture:
- 18.6
- 21.31
- 19.48
perimeter error:
- 2.497
- 5.486
- 3.094
radius error:
- 0.3961
- 0.7888
- 0.4743
smoothness error:
- 0.006953
- 0.004444
- 0.00624
symmetry error:
- 0.01782
- 0.01386
- 0.01397
texture error:
- 1.044
- 0.7975
- 0.7859
worst area:
- 677.9
- 1866
- 1156
worst compactness:
- 0.2378
- 0.2336
- 0.2394
worst concave points:
- 0.1015
- 0.1789
- 0.1514
worst concavity:
- 0.2671
- 0.2687
- 0.3791
worst fractal dimension:
- 0.0875
- 0.06589
- 0.08019
worst perimeter:
- 96.05
- 165.9
- 124.9
worst radius:
- 14.97
- 24.86
- 19.26
worst smoothness:
- 0.1426
- 0.1193
- 0.1546
worst symmetry:
- 0.3014
- 0.2551
- 0.2837
worst texture:
- 24.64
- 26.58
- 26
Model description
This is a HistGradientBoostingClassifier model trained on breast cancer dataset. It's trained with Halving Grid Search Cross Validation, with parameter grids on max_leaf_nodes and max_depth.
Intended uses & limitations
This model is not ready to be used in production.
Training Procedure
Hyperparameters
The model is trained with below hyperparameters.
Click to expand
| Hyperparameter | Value |
|---|---|
| aggressive_elimination | False |
| cv | 5 |
| error_score | nan |
| estimator__categorical_features | |
| estimator__early_stopping | auto |
| estimator__l2_regularization | 0.0 |
| estimator__learning_rate | 0.1 |
| estimator__loss | auto |
| estimator__max_bins | 255 |
| estimator__max_depth | |
| estimator__max_iter | 100 |
| estimator__max_leaf_nodes | 31 |
| estimator__min_samples_leaf | 20 |
| estimator__monotonic_cst | |
| estimator__n_iter_no_change | 10 |
| estimator__random_state | |
| estimator__scoring | loss |
| estimator__tol | 1e-07 |
| estimator__validation_fraction | 0.1 |
| estimator__verbose | 0 |
| estimator__warm_start | False |
| estimator | HistGradientBoostingClassifier() |
| factor | 3 |
| max_resources | auto |
| min_resources | exhaust |
| n_jobs | -1 |
| param_grid | {'max_leaf_nodes': [5, 10, 15], 'max_depth': [2, 5, 10]} |
| random_state | 42 |
| refit | True |
| resource | n_samples |
| return_train_score | True |
| scoring | |
| verbose | 0 |
Model Plot
The model plot is below.
HalvingGridSearchCV(estimator=HistGradientBoostingClassifier(), n_jobs=-1,param_grid={'max_depth': [2, 5, 10],'max_leaf_nodes': [5, 10, 15]},random_state=42)Please rerun this cell to show the HTML repr or trust the notebook.HalvingGridSearchCV(estimator=HistGradientBoostingClassifier(), n_jobs=-1,param_grid={'max_depth': [2, 5, 10],'max_leaf_nodes': [5, 10, 15]},random_state=42)HistGradientBoostingClassifier()
Evaluation Results
You can find the details about evaluation process and the evaluation results.
| Metric | Value |
|---|---|
| accuracy | 0.959064 |
| f1 score | 0.959064 |
How to Get Started with the Model
Use the code below to get started with the model.
Click to expand
import pickle
with open(pkl_filename, 'rb') as file:
clf = pickle.load(file)
Model Card Authors
This model card is written by following authors:
skops_user
Model Card Contact
You can contact the model card authors through following channels: [More Information Needed]
Citation
Below you can find information related to citation.
BibTeX:
bibtex
@inproceedings{...,year={2020}}
Additional Content
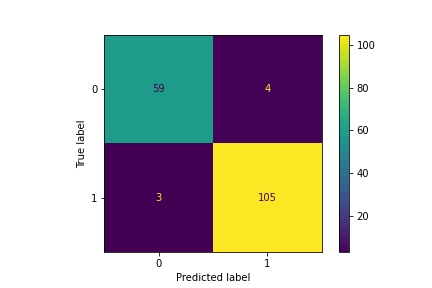
Confusion matrix
Hyperparameter search results
Click to expand
| iter | n_resources | mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_max_depth | param_max_leaf_nodes | params | split0_test_score | split1_test_score | split2_test_score | split3_test_score | split4_test_score | mean_test_score | std_test_score | rank_test_score | split0_train_score | split1_train_score | split2_train_score | split3_train_score | split4_train_score | mean_train_score | std_train_score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 44 | 0.0498069 | 0.0107112 | 0.0121156 | 0.0061838 | 2 | 5 | {'max_depth': 2, 'max_leaf_nodes': 5} | 0.875 | 0.5 | 0.625 | 0.75 | 0.375 | 0.625 | 0.176777 | 5 | 0.628571 | 0.628571 | 0.628571 | 0.514286 | 0.514286 | 0.582857 | 0.0559883 |
| 0 | 44 | 0.0492636 | 0.0187271 | 0.00738611 | 0.00245441 | 2 | 10 | {'max_depth': 2, 'max_leaf_nodes': 10} | 0.875 | 0.5 | 0.625 | 0.75 | 0.375 | 0.625 | 0.176777 | 5 | 0.628571 | 0.628571 | 0.628571 | 0.514286 | 0.514286 | 0.582857 | 0.0559883 |
| 0 | 44 | 0.0572055 | 0.0153176 | 0.0111395 | 0.0010297 | 2 | 15 | {'max_depth': 2, 'max_leaf_nodes': 15} | 0.875 | 0.5 | 0.625 | 0.75 | 0.375 | 0.625 | 0.176777 | 5 | 0.628571 | 0.628571 | 0.628571 | 0.514286 | 0.514286 | 0.582857 | 0.0559883 |
| 0 | 44 | 0.0498482 | 0.0177091 | 0.00857358 | 0.00415935 | 5 | 5 | {'max_depth': 5, 'max_leaf_nodes': 5} | 0.875 | 0.5 | 0.625 | 0.75 | 0.375 | 0.625 | 0.176777 | 5 | 0.628571 | 0.628571 | 0.628571 | 0.514286 | 0.514286 | 0.582857 | 0.0559883 |
| 0 | 44 | 0.0500658 | 0.00992094 | 0.00998321 | 0.00527031 | 5 | 10 | {'max_depth': 5, 'max_leaf_nodes': 10} | 0.875 | 0.5 | 0.625 | 0.75 | 0.375 | 0.625 | 0.176777 | 5 | 0.628571 | 0.628571 | 0.628571 | 0.514286 | 0.514286 | 0.582857 | 0.0559883 |
| 0 | 44 | 0.0525903 | 0.0151616 | 0.00874681 | 0.00462998 | 5 | 15 | {'max_depth': 5, 'max_leaf_nodes': 15} | 0.875 | 0.5 | 0.625 | 0.75 | 0.375 | 0.625 | 0.176777 | 5 | 0.628571 | 0.628571 | 0.628571 | 0.514286 | 0.514286 | 0.582857 | 0.0559883 |
| 0 | 44 | 0.0512018 | 0.0130152 | 0.00881834 | 0.00500514 | 10 | 5 | {'max_depth': 10, 'max_leaf_nodes': 5} | 0.875 | 0.5 | 0.625 | 0.75 | 0.375 | 0.625 | 0.176777 | 5 | 0.628571 | 0.628571 | 0.628571 | 0.514286 | 0.514286 | 0.582857 | 0.0559883 |
| 0 | 44 | 0.0566921 | 0.0186051 | 0.00513492 | 0.000498488 | 10 | 10 | {'max_depth': 10, 'max_leaf_nodes': 10} | 0.875 | 0.5 | 0.625 | 0.75 | 0.375 | 0.625 | 0.176777 | 5 | 0.628571 | 0.628571 | 0.628571 | 0.514286 | 0.514286 | 0.582857 | 0.0559883 |
| 0 | 44 | 0.060587 | 0.04041 | 0.00987453 | 0.00529624 | 10 | 15 | {'max_depth': 10, 'max_leaf_nodes': 15} | 0.875 | 0.5 | 0.625 | 0.75 | 0.375 | 0.625 | 0.176777 | 5 | 0.628571 | 0.628571 | 0.628571 | 0.514286 | 0.514286 | 0.582857 | 0.0559883 |
| 1 | 132 | 0.232459 | 0.0479878 | 0.0145514 | 0.00856422 | 10 | 5 | {'max_depth': 10, 'max_leaf_nodes': 5} | 0.961538 | 0.923077 | 0.923077 | 0.961538 | 0.961538 | 0.946154 | 0.0188422 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 1 | 132 | 0.272297 | 0.0228833 | 0.011561 | 0.0068272 | 10 | 10 | {'max_depth': 10, 'max_leaf_nodes': 10} | 0.961538 | 0.923077 | 0.923077 | 0.961538 | 0.961538 | 0.946154 | 0.0188422 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 1 | 132 | 0.239161 | 0.0330412 | 0.0116591 | 0.003554 | 10 | 15 | {'max_depth': 10, 'max_leaf_nodes': 15} | 0.961538 | 0.923077 | 0.923077 | 0.961538 | 0.961538 | 0.946154 | 0.0188422 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 2 | 396 | 0.920334 | 0.18198 | 0.0166654 | 0.00776263 | 10 | 15 | {'max_depth': 10, 'max_leaf_nodes': 15} | 0.962025 | 0.911392 | 0.987342 | 0.974359 | 0.935897 | 0.954203 | 0.0273257 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
Classification report
Click to expand
| index | precision | recall | f1-score | support |
|---|---|---|---|---|
| malignant | 0.951613 | 0.936508 | 0.944 | 63 |
| benign | 0.963303 | 0.972222 | 0.967742 | 108 |
| macro avg | 0.957458 | 0.954365 | 0.955871 | 171 |
| weighted avg | 0.958996 | 0.959064 | 0.958995 | 171 |