tags:
- monai
- medical
library_name: monai
license: apache-2.0
Description
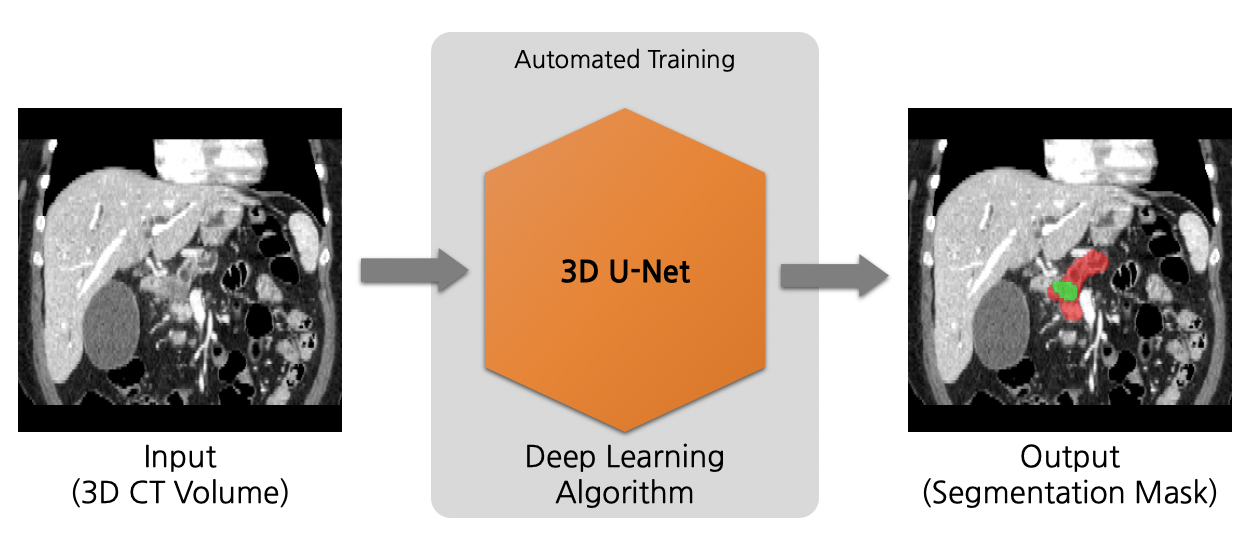
A neural architecture search algorithm for volumetric (3D) segmentation of the pancreas and pancreatic tumor from CT image.
Model Overview
This model is trained using the state-of-the-art algorithm [1] of the "Medical Segmentation Decathlon Challenge 2018" with 196 training images, 56 validation images, and 28 testing images.
This model is trained using the neural network model from the neural architecture search algorithm, DiNTS [1].
Data
The training dataset is Task07_Pancreas.tar from http://medicaldecathlon.com/. And the data list/split can be created with the script scripts/prepare_datalist.py.
Training configuration
The training was performed with at least 16GB-memory GPUs.
Actual Model Input: 96 x 96 x 96
Neural Architecture Search Configuration
The neural architecture search was performed with the following:
- AMP: True
- Optimizer: SGD
- Initial Learning Rate: 0.025
- Loss: DiceCELoss
Training Configuration
The training was performed with the following:
- AMP: True
- Optimizer: SGD
- (Initial) Learning Rate: 0.025
- Loss: DiceCELoss
- Note: If out-of-memory or program crash occurs while caching the data set, please change the cache_rate in CacheDataset to a lower value in the range (0, 1).
The segmentation of pancreas region is formulated as the voxel-wise 3-class classification. Each voxel is predicted as either foreground (pancreas body, tumour) or background. And the model is optimized with gradient descent method minimizing soft dice loss and cross-entropy loss between the predicted mask and ground truth segmentation.
Data Pre-processing and Augmentation
Input: 1 channel CT image with intensity in HU
- Converting to channel first
- Normalizing and clipping intensities of tissue window to [0,1]
- Cropping foreground surrounding regions
- Cropping random fixed sized regions of size [96, 96, 96] with the center being a foreground or background voxel at ratio 1 : 1
- Randomly rotating volumes
- Randomly zooming volumes
- Randomly smoothing volumes with Gaussian kernels
- Randomly scaling intensity of the volume
- Randomly shifting intensity of the volume
- Randomly adding Gaussian noises
- Randomly flipping volumes
Sliding-window Inference
Inference is performed in a sliding window manner with a specified stride.
Input and output formats
Input: 1 channel CT image
Output: 3 channels: Label 2: pancreatic tumor; Label 1: pancreas; Label 0: everything else
Performance
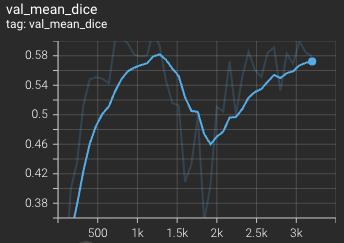
This model achieves the following Dice score on the validation data (our own split from the training dataset):
Mean Dice = 0.62
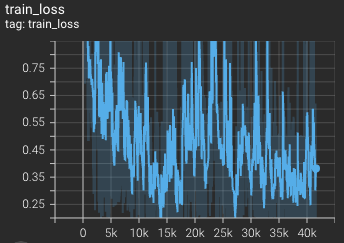
Training loss over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
Validation mean dice score over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
Searched Architecture Visualization
Users can install Graphviz for visualization of searched architectures (needed in custom/decode_plot.py). The edges between nodes indicate global structure, and numbers next to edges represent different operations in the cell searching space. An example of searched architecture is shown as follows:
Commands Example
Create data split (.json file):
python scripts/prepare_datalist.py --path /path-to-Task07_Pancreas/ --output configs/dataset_0.json
Execute model searching:
python -m scripts.search run --config_file configs/search.yaml
Execute multi-GPU model searching (recommended):
torchrun --nnodes=1 --nproc_per_node=8 -m scripts.search run --config_file configs/search.yaml
Execute training:
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.yaml --logging_file configs/logging.conf
Override the train config to execute multi-GPU training:
torchrun --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.yaml','configs/multi_gpu_train.yaml']" --logging_file configs/logging.conf
Override the train config to execute evaluation with the trained model:
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.yaml','configs/evaluate.yaml']" --logging_file configs/logging.conf
Execute inference:
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.yaml --logging_file configs/logging.conf
Export checkpoint for TorchScript
python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.yaml
Disclaimer
This is an example, not to be used for diagnostic purposes.
References
[1] He, Y., Yang, D., Roth, H., Zhao, C. and Xu, D., 2021. Dints: Differentiable neural network topology search for 3d medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 5841-5850).
License
Copyright (c) MONAI Consortium
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.