|
--- |
|
tags: |
|
- monai |
|
- medical |
|
library_name: monai |
|
license: apache-2.0 |
|
--- |
|
# Description |
|
A neural architecture search algorithm for volumetric (3D) segmentation of the pancreas and pancreatic tumor from CT image. |
|
|
|
# Model Overview |
|
This model is trained using the state-of-the-art algorithm [1] of the "Medical Segmentation Decathlon Challenge 2018" with 196 training images, 56 validation images, and 28 testing images. |
|
|
|
This model is trained using the neural network model from the neural architecture search algorithm, DiNTS [1]. |
|
|
|
 |
|
|
|
## Data |
|
The training dataset is Task07_Pancreas.tar from http://medicaldecathlon.com/. And the data list/split can be created with the script `scripts/prepare_datalist.py`. |
|
|
|
## Training configuration |
|
The training was performed with at least 16GB-memory GPUs. |
|
|
|
Actual Model Input: 96 x 96 x 96 |
|
|
|
### Neural Architecture Search Configuration |
|
The neural architecture search was performed with the following: |
|
|
|
- AMP: True |
|
- Optimizer: SGD |
|
- Initial Learning Rate: 0.025 |
|
- Loss: DiceCELoss |
|
|
|
### Training Configuration |
|
The training was performed with the following: |
|
|
|
- AMP: True |
|
- Optimizer: SGD |
|
- (Initial) Learning Rate: 0.025 |
|
- Loss: DiceCELoss |
|
- Note: If out-of-memory or program crash occurs while caching the data set, please change the cache\_rate in CacheDataset to a lower value in the range (0, 1). |
|
|
|
The segmentation of pancreas region is formulated as the voxel-wise 3-class classification. Each voxel is predicted as either foreground (pancreas body, tumour) or background. And the model is optimized with gradient descent method minimizing soft dice loss and cross-entropy loss between the predicted mask and ground truth segmentation. |
|
|
|
### Data Pre-processing and Augmentation |
|
|
|
Input: 1 channel CT image with intensity in HU |
|
|
|
- Converting to channel first |
|
- Normalizing and clipping intensities of tissue window to [0,1] |
|
- Cropping foreground surrounding regions |
|
- Cropping random fixed sized regions of size [96, 96, 96] with the center being a foreground or background voxel at ratio 1 : 1 |
|
- Randomly rotating volumes |
|
- Randomly zooming volumes |
|
- Randomly smoothing volumes with Gaussian kernels |
|
- Randomly scaling intensity of the volume |
|
- Randomly shifting intensity of the volume |
|
- Randomly adding Gaussian noises |
|
- Randomly flipping volumes |
|
|
|
### Sliding-window Inference |
|
Inference is performed in a sliding window manner with a specified stride. |
|
|
|
## Input and output formats |
|
Input: 1 channel CT image |
|
|
|
Output: 3 channels: Label 2: pancreatic tumor; Label 1: pancreas; Label 0: everything else |
|
|
|
## Performance |
|
This model achieves the following Dice score on the validation data (our own split from the training dataset): |
|
|
|
Mean Dice = 0.62 |
|
|
|
Training loss over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve) |
|
|
|
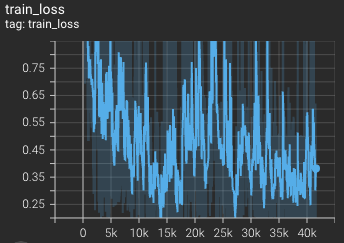 |
|
|
|
Validation mean dice score over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve) |
|
|
|
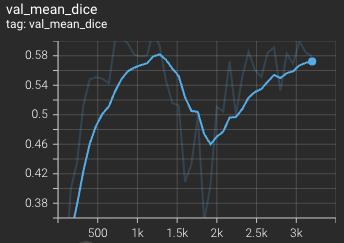 |
|
|
|
### Searched Architecture Visualization |
|
Users can install Graphviz for visualization of searched architectures (needed in custom/decode_plot.py). The edges between nodes indicate global structure, and numbers next to edges represent different operations in the cell searching space. An example of searched architecture is shown as follows: |
|
|
|
## Commands Example |
|
Create data split (.json file): |
|
|
|
``` |
|
python scripts/prepare_datalist.py --path /path-to-Task07_Pancreas/ --output configs/dataset_0.json |
|
``` |
|
|
|
Execute model searching: |
|
|
|
``` |
|
python -m scripts.search run --config_file configs/search.yaml |
|
``` |
|
|
|
Execute multi-GPU model searching (recommended): |
|
|
|
``` |
|
torchrun --nnodes=1 --nproc_per_node=8 -m scripts.search run --config_file configs/search.yaml |
|
``` |
|
|
|
Execute training: |
|
|
|
``` |
|
python -m monai.bundle run training --meta_file configs/metadata.json --config_file configs/train.yaml --logging_file configs/logging.conf |
|
``` |
|
|
|
Override the `train` config to execute multi-GPU training: |
|
|
|
``` |
|
torchrun --nnodes=1 --nproc_per_node=2 -m monai.bundle run training --meta_file configs/metadata.json --config_file "['configs/train.yaml','configs/multi_gpu_train.yaml']" --logging_file configs/logging.conf |
|
``` |
|
|
|
Override the `train` config to execute evaluation with the trained model: |
|
|
|
``` |
|
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file "['configs/train.yaml','configs/evaluate.yaml']" --logging_file configs/logging.conf |
|
``` |
|
|
|
Execute inference: |
|
|
|
``` |
|
python -m monai.bundle run evaluating --meta_file configs/metadata.json --config_file configs/inference.yaml --logging_file configs/logging.conf |
|
``` |
|
|
|
Export checkpoint for TorchScript |
|
|
|
``` |
|
python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.yaml |
|
``` |
|
|
|
# Disclaimer |
|
This is an example, not to be used for diagnostic purposes. |
|
|
|
# References |
|
[1] He, Y., Yang, D., Roth, H., Zhao, C. and Xu, D., 2021. Dints: Differentiable neural network topology search for 3d medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 5841-5850). |
|
|
|
# License |
|
Copyright (c) MONAI Consortium |
|
|
|
Licensed under the Apache License, Version 2.0 (the "License"); |
|
you may not use this file except in compliance with the License. |
|
You may obtain a copy of the License at |
|
|
|
http://www.apache.org/licenses/LICENSE-2.0 |
|
|
|
Unless required by applicable law or agreed to in writing, software |
|
distributed under the License is distributed on an "AS IS" BASIS, |
|
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. |
|
See the License for the specific language governing permissions and |
|
limitations under the License. |
|
|