licenses
sequencelengths 1
3
| version
stringclasses 677
values | tree_hash
stringlengths 40
40
| path
stringclasses 1
value | type
stringclasses 2
values | size
stringlengths 2
8
| text
stringlengths 25
67.1M
| package_name
stringlengths 2
41
| repo
stringlengths 33
86
|
|---|---|---|---|---|---|---|---|---|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 3822 | using REPL
abstract type CharStyle end
struct Block <: CharStyle end
struct Braile <: CharStyle end
"""
REPLOutput <: GraphicOutput
REPLOutput(init; tspan, kw...)
An output that is displayed directly in the REPL. It can either store or discard
simulation frames.
# Arguments:
- `init`: initialisation `AbstractArrayArray` or `NamedTuple` of `AbstractArrayArray`.
# Keywords
- `color`: a color from Crayons.jl
- `cutoff`: `Real` cutoff point to display a full or empty cell. Default is `0.5`
- `style`: `CharStyle` `Block()` or `Braile()` printing. `Braile` uses 1/4 the screen space of `Block`.
$GRAPHICOUTPUT_KEYWORDS
e `GraphicConfig` object can be also passed to the `graphicconfig` keyword, and other keywords will be ignored.
"""
mutable struct REPLOutput{T,F<:AbstractVector{T},E,GC,Co,St,Cu} <: GraphicOutput{T,F}
frames::F
running::Bool
extent::E
graphicconfig::GC
color::Co
style::St
cutoff::Cu
end
function REPLOutput(;
frames, running, extent, graphicconfig,
color=:white, cutoff=0.5, style=Block(), kw...
)
if store(graphicconfig)
append!(frames, _zerogrids(init(extent), length(tspan(extent))-1))
end
REPLOutput(frames, running, extent, graphicconfig, color, style, cutoff)
end
function showframe(frame::AbstractArray, o::REPLOutput, data::AbstractSimData)
_print_to_repl((0, 0), o.color, _replframe(o, frame, currentframe(data)))
# Print the timestamp in the top right corner
_print_to_repl((0, 0), o.color, string("Time $(currenttime(data))"))
end
# Terminal commands
_savepos(io::IO=terminal.out_stream) = print(io, "\x1b[s")
_restorepos(io::IO=terminal.out_stream) = print(io, "\x1b[u")
_movepos(io::IO, c=(0,0)) = print(io, "\x1b[$(c[2]);$(c[1])H")
_cursor_hide(io::IO=terminal.out_stream) = print(io, "\x1b[?25l")
_cursor_show(io::IO=terminal.out_stream) = print(io, "\x1b[?25h")
_print_to_repl(pos, c::Symbol, s::String) = _print_to_repl(pos, Crayon(foreground=c), s)
function _print_to_repl(pos, color::Crayon, str::String)
io = terminal.out_stream
_savepos(io)
_cursor_hide(io)
_movepos(io, pos)
print(io, color)
print(io, str)
_cursor_show(io)
_restorepos(io)
end
# Block size constants to calculate the frame size as
# braile pixels are half the height and width of block pixels
const YBRAILE = 4
const XBRAILE = 2
const YBLOCK = 2
const XBLOCK = 1
_chartype(o::REPLOutput) = _chartype(o.style)
_chartype(s::Braile) = YBRAILE, XBRAILE, brailize
_chartype(s::Block) = YBLOCK, XBLOCK, blockize
function _replframe(o, frame::AbstractArray{<:Any,N}, currentframe) where N
ystep, xstep, charfunc = _chartype(o)
# Limit output area to available terminal size.
dispy, dispx = displaysize(stdout)
if N === 1
offset = 0
rnge = max(1, xstep * offset):min(length(frame))
f = currentframe
nrows = min(f, dispy)
# For 1D we show all the rows every time
tlen = length(tspan(o))
rowstrings = map(f - nrows + 1:f) do i
framewindow1 = view(adapt(Array, frames(o)[i]), rnge)
framewindow2 = if i == tlen
framewindow1
else
view(adapt(Array, frames(o)[i]), rnge)
end
charfunc(PermutedDimsArray(hcat(framewindow1, framewindow2), (2, 1)), o.cutoff)
end
return join(rowstrings, "\n")
else
youtput, xoutput = outputsize = size(frame)
yoffset, xoffset = (0, 0)
yrange = max(1, ystep * yoffset):min(youtput, ystep * (dispy + yoffset - 1))
xrange = max(1, xstep * xoffset):min(xoutput, xstep * (dispx + xoffset - 1))
framewindow = view(adapt(Array, frame), yrange, xrange) # TODO make this more efficient on GPU
return charfunc(framewindow, o.cutoff)
end
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 875 | """
Greyscale
Greyscale(min=nothing, max=nothing)
A greeyscale scheme ith better performance than using a
Colorschemes.jl scheme as there is not array access or interpolation.
`min` and `max` are values between `0.0` and `1.0` that define the range of greys used.
"""
struct Greyscale{M1,M2}
min::M1
max::M2
end
Greyscale(; min=nothing, max=nothing) = Greyscale(min, max)
Base.get(scheme::Greyscale, x::Real) = scale(x, scheme.min, scheme.max)
const Grayscale = Greyscale
"""
ObjectScheme
ObjectScheme()
Default colorscheme. Similar to `GreyScale` for `Number`.
Other grid objects can define a custom method to return colors from composite objects:
```julia
DynamicGrids.to_rgb(::ObjectScheme, obj::MyObjectType) = ...
```
Which must return an `ARGB32` value.
"""
struct ObjectScheme end
to_rgb(scheme::ObjectScheme, x::Real) = to_rgb(x)
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 3334 | """
TextConfig
TextConfig(; kw...)
TextConfig(face, namepixels, namepos, timepixels, timepos, fcolor, bcolor)
Text configuration for printing timestep and grid name on the image.
# Arguments / Keywords
- `font`: A `FreeTypeAbstraction.FTFont`, or a `String` with the font name to look for. The `FTFont` may load more quickly.
- `namepixels` and `timepixels`: the pixel size of the font.
- `timepos` and `namepos`: tuples that set the label positions, in `Int` pixels.
- `fcolor` and `bcolor`: the foreground and background colors, as `ARGB32`.
"""
struct TextConfig{F,NPi,NPo,TPi,TPo,FC,BC}
face::F
namepixels::NPi
namepos::NPo
timepixels::TPi
timepos::TPo
fcolor::FC
bcolor::BC
end
function TextConfig(;
font=autofont(), namepixels=12, timepixels=12,
namepos=(3timepixels + namepixels, timepixels),
timepos=(2timepixels, timepixels),
fcolor=ARGB32(1.0), bcolor=ZEROCOL,
)
if font isa FreeTypeAbstraction.FTFont
face = font
elseif font isa AbstractString
face = FreeTypeAbstraction.findfont(font)
face isa Nothing && _fontnotfounderror(font)
else
_fontnotstring(font)
end
TextConfig(face, namepixels, namepos, timepixels, timepos, fcolor, bcolor)
end
function autofont()
fonts = if Sys.islinux()
("cantarell", "sans-serif", "Bookman")
else
("arial", "sans-serif")
end
for font in fonts
face = FreeTypeAbstraction.findfont(font)
face isa Nothing || return font
end
_nodefaultfonterror(fonts)
end
@noinline _fontnotstring(font) = throw(ArgumentError("font $font is not a String"))
@noinline _fontnotfounderror(font) =
throw(ArgumentError(
"""
Font "$font" wasn't be found in this system. Specify an existing font name
with the `font` keyword, or use `text=nothing` to display no text."
"""
))
@noinline _nodefaultfonterror(font) =
error(
"""
Your system does not contain the default font $font. Specify an existing font
name `String` with the keyword-argument `font`, for the `Output` or `ImageConfig`.
"""
)
# Render time `name` and `t` as text onto the image, following config settings.
function _rendertime! end
function _rendertext!(img, config::TextConfig, name, t)
_rendername!(img, config::TextConfig, name)
_rendertime!(img, config::TextConfig, t)
img
end
_rendertext!(img, config::Nothing, name, t) = nothing
# Render `name` as text on the image following config settings.
function _rendername!(img, config::TextConfig, name)
renderstring!(img, name, config.face, config.namepixels, config.namepos...;
fcolor=config.fcolor, bcolor=config.bcolor
)
img
end
_rendername!(img, config::TextConfig, name::Nothing) = img
_rendername!(img, config::Nothing, name) = img
_rendername!(img, config::Nothing, name::Nothing) = img
# Render time `t` as text on the image following config settings.
function _rendertime!(img, config::TextConfig, t)
renderstring!(img, string(t), config.face, config.timepixels, config.timepos...;
fcolor=config.fcolor, bcolor=config.bcolor
)
img
end
_rendertime!(img, config::Nothing, t) = img
_rendertime!(img, config::TextConfig, t::Nothing) = img
_rendertime!(img, config::Nothing, t::Nothing) = img
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 3304 | """
TransformedOutput(f, init; tspan::AbstractRange, kw...)
An output that stores the result of some function `f` of the grid/s.
# Arguments
- `f`: a function or functor that accepts an `AbstractArray` or `NamedTuple` of
`AbstractArray` with names matching `init`. The `AbstractArray` will be a view into
the grid the same size as the init grids, removing any padding that has been added.
- `init`: initialisation `Array` or `NamedTuple` of `Array`
# Keywords
- `tspan`: `AbstractRange` timespan for the simulation
- `aux`: NamedTuple of arbitrary input data. Use `get(data, Aux(:key), I...)`
to access from a `Rule` in a type-stable way.
- `mask`: `BitArray` for defining cells that will/will not be run.
- `padval`: padding value for grids with neighborhood rules. The default is `zero(eltype(init))`.
$EXPERIMENTAL
"""
mutable struct TransformedOutput{T,A<:AbstractVector{T},E,F,B} <: Output{T,A}
frames::A
running::Bool
extent::E
f::F
buffer::B
end
function TransformedOutput(f::Function, init::Union{NamedTuple,AbstractMatrix}; extent=nothing, kw...)
# We have to handle some things manually as we are changing the standard output frames
extent = extent isa Nothing ? Extent(; init=init, kw...) : extent
# Define buffers to copy to before applying `f`
buffer = init isa NamedTuple ? map(zero, init) : zero(init)
zeroframe = f(buffer)
# Build simulation frames from the output of `f` for empty frames
frames = [deepcopy(zeroframe) for f in eachindex(tspan(extent))]
# Set the first frame to the output of `f` for `init`
frames[1] = f(init)
return TransformedOutput(frames, false, extent, f, buffer)
end
function TransformedOutput(init; kw...)
throw(ArgumentError("TransformedOutput must be passed a function and the init grid(s) as arguments"))
end
function storeframe!(o::TransformedOutput, data::AbstractSimData)
transformed = _transform_grids(o, grids(data))
i = frameindex(o, data)
# Copy the transformed grid/s to the output frames,
# instead of just assigning (see issue #169)
o[i] = _copytransformed!(o[i], transformed)
end
# Copy arrays manually as reducing functions can return the original object without copy.
_copytransformed!(dest::NamedTuple, src::NamedTuple) = map(_copytransformed!, dest, src)
_copytransformed!(dest::AbstractArray, src::AbstractArray) = dest .= src
# Non-array output is just assigned
_copytransformed!(dest, src) = src
# Multi/named grid simulation, f is passed a NamedTuple
function _transform_grids(o::TransformedOutput, grids::NamedTuple)
# Make a new named tuple of raw arrays without wrappers, copying
# to the buffer where an OffsetArray was used for padding.
# Often it's faster to copy than use a view when f is sum/mean etc.
nt = map(grids, o.buffer) do g, b
source(g) isa OffsetArray ? copy!(b, sourceview(g)) : source(g)
end
o.f(nt)
end
# Single unnamed grid simulation, f is passed an AbstractArray
function _transform_grids(o::TransformedOutput, grids::NamedTuple{(DEFAULT_KEY,)})
g = first(grids)
A = source(g) isa OffsetArray ? copy!(o.buffer, sourceview(g)) : source(g)
o.f(A)
end
init_output_grids!(o::TransformedOutput, init) = nothing
initdata!(o::TransformedOutput, init) = nothing
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 796 | using DynamicGrids, Test, Adapt
using DynamicGrids: SimData
output = ArrayOutput(BitArray(rand(Bool, 10, 10));
tspan=1:10,
mask=BitArray(rand(Bool, 10, 10)),
aux=(aux1=BitArray(rand(Bool, 10, 10)),),
)
rs1 = Ruleset(Life(); opt=NoOpt())
rs2 = Ruleset(Life(); opt=SparseOpt())
sd1 = SimData(output, rs1)
sd2 = SimData(output, rs2)
Adapt.adapt(Array, rs1)
Adapt.adapt(Array, rs2)
b_sd1 = Adapt.adapt(Array, sd1)
b_sd2 = Adapt.adapt(Array, sd2)
@test b_sd2.extent.init._default_ isa Array
@test b_sd2.extent.mask isa Array
@test b_sd2.extent.aux.aux1 isa Array
@test parent(b_sd2.grids[:_default_].source) isa Array
@test parent(b_sd2.grids[:_default_].dest) isa Array
@test parent(b_sd2.grids[:_default_].optdata.sourcestatus) isa Array
@test Adapt.adapt(Array, output)[1] isa Array
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 4588 | using DynamicGrids, Test, BenchmarkTools
using DynamicGrids: SimData, radius, rules, _readkeys, _writekeys,
applyrule, neighborhood, neighborhoodkey, Extent, ruletype
@testset "CellRule chain" begin
rule1 = Cell{:a,:b}() do data, a, I
2a
end
rule2 = Cell{Tuple{:b,:d},:c}() do data, (b, d), I
b + d
end
rule3 = Cell{Tuple{:a,:c,:d},Tuple{:d,:e}}() do data, (a, c, d), I
a + c + d, 3a
end
rule4 = Cell{Tuple{:a,:b,:c,:d},Tuple{:a,:b,:c,:d}}() do data, (a, b, c, d), I
2a, 2b, 2c, 2d
end
# These aren't actually used yet, they just build SimData
agrid = [1 0 0
0 0 2]
bgrid = [0 0 0
0 0 0]
cgrid = [0 0 0
0 0 0]
dgrid = [0 0 0
0 0 0]
egrid = [0 0 0
0 0 0]
chain = Chain(rule1, rule2, rule3, rule4)
@test ruletype(chain) == CellRule
@test _readkeys(chain) == (:a, :b, :d, :c)
@test _writekeys(chain) == (:b, :c, :d, :e, :a)
ruleset = Ruleset(chain)
init = (a=agrid, b=bgrid, c=cgrid, d=dgrid, e=egrid)
data = SimData(Extent(init=init, tspan=1:1), ruleset)
@test radius(ruleset) == (b=0, c=0, d=0, e=0, a=0)
@test applyrule(data, chain, (b=1, c=1, d=1, a=1), (1, 1)) ==
(4, 6, 10, 3, 2)
# @inferred applyrule(data, chain, (b=1, c=1, d=1, a=1), (1, 1))
state = (b=1, c=1, d=1, a=1)
ind = (1, 1)
# This breaks with --inline=no
# b = @benchmark applyrule($data, $chain, $state, $ind)
# @test b.allocs == 0
output = ArrayOutput(init; tspan=1:3)
sim!(output, ruleset)
@test output[2][:a] == [2 0 0
0 0 4]
@test output[3][:a] == [4 0 0
0 0 8]
@test output[2][:b] == [4 0 0
0 0 8]
@test output[3][:b] == [8 0 0
0 0 16]
@test output[2][:c] == [4 0 0
0 0 8]
@test output[3][:c] == [20 0 0
0 0 40]
@test output[3][:d] == [36 0 0
0 0 72]
@test output[3][:d] == [36 0 0
0 0 72]
@test output[2][:e] == [3 0 0
0 0 6]
@test output[3][:e] == [6 0 0
0 0 12]
@test isinferred(output, ruleset)
end
@testset "NeighborhoodRule, CellRule chain" begin
buf = reshape(1:9, 3, 3)
hood = Moore{1}(buf)
hoodrule = Neighbors{:a,:a}(hood) do data, neighborhodhood, cell, I
sum(neighborhodhood)
end
rule = Cell{Tuple{:a,:c},:b}() do data, (b, c), I
b + c
end
init = (
a = [0 0 0 0 0
0 0 0 0 0
0 0 1 0 0
0 0 0 0 0
0 0 0 0 0],
b = [0 0 0 0 0
0 0 0 0 0
0 0 0 0 0
0 0 0 0 0
0 0 0 0 0],
c = [1 1 1 1 1
1 1 1 1 1
1 1 1 1 1
1 1 1 1 1
1 1 1 1 1]
)
chain = Chain(hoodrule, rule)
@test radius(chain) === 1
@test ruletype(chain) == NeighborhoodRule
@test neighborhood(chain) == hood
@test Tuple(neighbors(chain)) === (1, 2, 3, 4, 6, 7, 8, 9)
@test neighborhoodkey(chain) === :a
@test rules(Base.tail(chain)) === (rule,)
@test chain[1] === first(chain) === hoodrule
@test chain[end] === last(chain) === rule
@test length(chain) === 2
@test iterate(chain) === (hoodrule, 2)
@test firstindex(chain) === 1
@test lastindex(chain) === 2
ruleset = Ruleset(chain; opt=NoOpt())
noopt_output = ArrayOutput(init; tspan=1:3)
@btime sim!($noopt_output, $ruleset)
@test isinferred(noopt_output, ruleset)
ruleset = Ruleset(Chain(hoodrule, rule); opt=SparseOpt())
sparseopt_output = ArrayOutput(init; tspan=1:3)
@btime sim!($sparseopt_output, $ruleset; init=$init)
@test isinferred(sparseopt_output, ruleset)
noopt_output[2][:a] == sparseopt_output[2][:a] ==
[0 0 0 0 0
0 1 1 1 0
0 1 0 1 0
0 1 1 1 0
0 0 0 0 0]
noopt_output[2][:b] == sparseopt_output[2][:b] ==
[1 1 1 1 1
1 2 2 2 1
1 2 1 2 1
1 2 2 2 1
1 1 1 1 1]
noopt_output[3][:a] == sparseopt_output[3][:a] ==
[1 2 3 2 1
2 2 4 2 2
3 4 8 4 3
2 2 4 2 2
1 2 3 2 1]
noopt_output[3][:b] == sparseopt_output[3][:b] ==
[2 3 4 3 2
3 3 5 3 3
4 5 9 5 4
3 3 5 3 3
2 3 4 3 2]
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 2556 | using DynamicGrids, Test, Dates
using DynamicGrids: ruletype
@testset "RunIf" begin
@testset "CellRule" begin
rule = Cell{:a,:a}() do data, a, I
10a
end
condition = RunIf(rule) do data, state, index
state < 3oneunit(state)
end
init = (a=[1 2 3; 0 4 -1],)
@test radius(condition) == (a=0)
@test ruletype(condition) == CellRule
@test ruletype(condition) == CellRule
output = ArrayOutput(init; tspan=1:3)
sim!(output, condition)
@test output[2][:a] == [10 20 3; 0 4 -10]
@test output[3][:a] == [10 20 3; 0 4 -100]
end
@testset "NeighborhoodRule" begin
neighborsrule = Neighbors{:a,:a}(Moore{1}()) do data, hood, a, I
sum(hood)
end
condition = RunIf(neighborsrule) do data, state, index
state == 1
end
init = (a=[0 0 0 0;
0 1 0 0;
1 0 1 0;
0 0 0 1],)
@test radius(condition) == (a=1)
@test ruletype(condition) == NeighborhoodRule
output = ArrayOutput(init; tspan=1:3)
sim!(output, condition)
@test output[2][:a] == [0 0 0 0;
0 2 0 0;
1 0 2 0;
0 0 0 1]
@test output[3][:a] == [0 0 0 0;
0 2 0 0;
2 0 2 0;
0 0 0 2]
# @test isinferred(output, condition)
end
end
@testset "RunAt" begin
rule = Cell{:a,:a}((d, a, I) -> a + 1)
timedrule1 = Cell{:a,:a}((d, a, I) -> 4a)
timedrule2 = Cell{:a,:a}((d, a, I) -> a ÷ 2)
runatrule = RunAt(timedrule1, timedrule2; times=DateTime(2001, 3):Month(2):DateTime(2001, 5))
init = (a=[1 2 3; 0 4 -5],)
@test radius(runatrule) == 0
@test length(runatrule) == 2
@test runatrule[1] === timedrule1
@test runatrule[2] === timedrule2
@test Tuple(rule for rule in runatrule) == rules(runatrule)
@test Base.tail(runatrule) == RunAt(timedrule2; times=DateTime(2001, 3):Month(2):DateTime(2001, 5))
@test firstindex(runatrule) === 1
@test lastindex(runatrule) === 2
output = ArrayOutput(init; tspan=DateTime(2001,1):Month(1):DateTime(2001,5))
sim!(output, rule, runatrule)
@test output[2][:a] == [2 3 4; 1 5 -4]
@test output[3][:a] == [6 8 10; 4 12 -6]
@test output[4][:a] == [7 9 11; 5 13 -5]
@test output[5][:a] == [16 20 24; 12 28 -8]
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 2262 | using DynamicGrids, Test
using DynamicGrids: SimData, WritableGridData, Extent,
_getreadgrids, _getwritegrids, _readcell, _writecell!, grids, source, dest
rule = Cell{Tuple{:c,:a,:b},Tuple{:a,:c}}(identity)
init = (a=fill(1, 4, 4), b=fill(2, 4, 4), c=fill(3, 4, 4))
simdata = SimData(Extent(;init=init, tspan=1:1), Ruleset(rule))
@testset "_getreadgrids gets read grids for a Rule" begin
rkeys, rgrids = _getreadgrids(rule, simdata)
@test rkeys === (Val(:c), Val(:a), Val(:b))
@test rgrids === (simdata[:c], simdata[:a], simdata[:b])
end
@testset "_getwritegrids gets write grids for a Rule" begin
wkeys, wgrids = _getwritegrids(rule, simdata)
@test wkeys === (Val(:a), Val(:c))
@test wgrids === map(WritableGridData, (simdata[:a], simdata[:c]))
end
@testset "_readcell read from specified grids" begin
@test _readcell(simdata, Val(:a), 1, 1) == 1
@test _readcell(simdata, Val(:b), 1, 1) == 2
@test _readcell(simdata, Val(:c), 1, 1) == 3
@test _readcell(simdata, (Val(:c), Val(:a), Val(:b)), 1, 1) == (c=3, a=1, b=2)
@test _readcell(simdata, (Val(:a), Val(:c)), 1, 1) == (a=1, c=3)
end
@testset "_writecell writes to source for CellRule" begin
simdata = SimData(Extent(;init=init, tspan=1:1), Ruleset(rule))
_writecell!(simdata, Val(CellRule), (Val(:c), Val(:a), Val(:b)), (8, 6, 7), 1, 2)
@test map(g -> source(g)[1, 2], grids(simdata)) == (a=6, b=7, c=8)
@test map(g -> dest(g)[1, 2], grids(simdata)) == (a=1, b=2, c=3)
simdata = SimData(Extent(;init=init, tspan=1:1), Ruleset(rule))
_writecell!(simdata, Val(CellRule), Val(:c), 99, 4, 3)
@test source(simdata[:c])[4, 3] == 99
@test dest(simdata[:c])[4, 3] == 3
end
@testset "_writecell writes to dest for other Rules" begin
simdata = SimData(Extent(;init=init, tspan=1:1), Ruleset(rule))
_writecell!(simdata, Val(Rule), (Val(:b), Val(:a)), (11, 10), 4, 4)
@test map(g -> source(g)[4, 4], grids(simdata)) == (a=1, b=2, c=3)
@test map(g -> dest(g)[4, 4], grids(simdata)) == (a=10, b=11, c=3)
simdata = SimData(Extent(;init=init, tspan=1:1), Ruleset(rule))
_writecell!(simdata, Val(Rule), Val(:c), 99, 4, 3)
@test source(simdata[:c])[4, 3] == 3
@test dest(simdata[:c])[4, 3] == 99
end
nothing
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 11995 | using DynamicGrids, Dates, Test, Colors, ColorSchemes, FileIO
using FreeTypeAbstraction
using DynamicGrids: render!, renderer, minval, maxval, normalise, SimData, NoDisplayImageOutput,
isstored, isasync, initialise!, finalise!, maybesleep, fps, settimestamp!, timestamp, textconfig,
tspan, setfps!, frames, isshowable, showframe, to_rgb, scale, Extent, extent,
_autokeys, _autolayout
using ColorSchemes: leonardo
@testset "to_rgb" begin
@test to_rgb(0.5) === ARGB32(0.5, 0.5, 0.5, 1.0)
@test to_rgb((0.5, 0.5, 0.5)) === ARGB32(0.5, 0.5, 0.5, 1.0)
@test to_rgb((0.5, 0.5, 0.5, 1.0)) === ARGB32(0.5, 0.5, 0.5, 1.0)
@test to_rgb(RGB(0.5, 0.5, 0.5)) === ARGB32(0.5, 0.5, 0.5, 1.0)
@test to_rgb(ARGB32(0.5, 0.5, 0.5)) === ARGB32(0.5, 0.5, 0.5, 1.0)
@test to_rgb(Greyscale(), 0.5) === ARGB32(0.5, 0.5, 0.5, 1.0)
@test to_rgb(ObjectScheme(), 0.5) === ARGB32(0.5, 0.5, 0.5, 1.0)
end
@testset "normalise" begin
@test normalise(-.2, 0.0, 1.0) == 0.0
@test normalise(1.2, 0.0, 1.0) == 1.0
@test normalise(-.2, 0.0, nothing) == 0.0
@test normalise(1.2, nothing, 1.0) == 1.0
@test normalise(1.2, nothing, nothing) == 1.2
end
@testset "scale" begin
@test scale(0.0, 5.0, 10.0) == 5.0
@test scale(0.5, 5.0, 10.0) == 7.5
@test scale(1.0, nothing, 10.0) == 10.0
@test scale(0.0, -2.0, nothing) == -2.0
@test scale(1.2, nothing, nothing) == 1.2
end
l0 = ARGB32(get(leonardo, 0))
l05 = ARGB32(get(leonardo, 0.5))
l08 = ARGB32(get(leonardo, 0.8))
l1 = ARGB32(get(leonardo, 1))
images = []
DynamicGrids.showimage(image, o::NoDisplayImageOutput) = begin
push!(images, image)
image
end
@testset "basic ImageOutput" begin
init_ = [8.0 10.0;
NaN 5.0]
output = NoDisplayImageOutput(init_; tspan=1:1, maxval=40.0, text=nothing)
@test all(parent(output)[1] .=== init_)
@test minval(output) === 0
@test maxval(output) === 40.0
@test textconfig(output) === nothing
@test renderer(output).scheme == ObjectScheme()
@test isasync(output) == false
@test isstored(output) == false
@test maybesleep(output, 1.0) === nothing
@test timestamp(output) === 0.0
pre = time()
settimestamp!(output, 1)
@test timestamp(output) > pre
@test length(output) == 1
push!(output, 2init_)
@test length(output) == 2
@test all(output[2] .=== 2init_)
@test tspan(output) == 1:1
@test fps(output) === 25.0
@test setfps!(output, 1000.0) === 1000.0
@test fps(output) === 1000.0
output[1] = 5init_
@test all(frames(output)[1] .=== 5init_)
@test isshowable(output, 1)
rndr = Image()
output = NoDisplayImageOutput(init_;
tspan=1:10, maxval=40.0, renderer=rndr, text=nothing
)
simdata = SimData(output, Ruleset(Life()))
z0 = DynamicGrids.ZEROCOL
ref = [ARGB32(0.2, 0.2, 0.2) ARGB32(0.25, 0.25, 0.25)
z0 ARGB32(0.125, 0.125, 0.125)]
@test showframe(output, simdata) == ref
savegif("test.gif", output)
gif = load("test.gif")
@test gif == RGB.(ref)
rm("test.gif")
end
@testset "Renderer" begin
@testset "auto render layout" begin
@test _autokeys((a=[0], b=[0], c=[0])) == (:a, :b, :c)
@test _autokeys((a=[[0,0]], b=[1], c=[[3,4]])) == (:a=>1, :a=>2, :b, :c=>1, :c=>2)
@test _autolayout([0]) == reshape(Any[1], 1, 1)
@test _autolayout((a=[0], b=[0], c=[0])) == Any[:a :b :c]
@testset "Empty layout cells are filled with nothing" begin
@test _autolayout((a=[[0,0]], b=[1], c=[[3,4]])) == Any[:a=>1 :b :c=>2; :a=>2 :c=>1 nothing]
end
end
init_ = [8.0 10.0;
0.0 5.0]
mask_ = Bool[0 1;
1 1]
rndr = Image(zerocolor=(1.0, 0.0, 0.0), maskcolor=(0.1, 0.1, 0.1))
ic = DynamicGrids.ImageConfig(init_; renderer=rndr, textconfig=nothing)
@test ic.renderer === rndr
output = NoDisplayImageOutput((a=init_,);
tspan=DateTime(2001):Year(1):DateTime(2010), mask=mask_,
renderer=rndr, text=nothing, minval=0.0, maxval=10.0, store=true
)
@test renderer(output) === output.imageconfig.renderer === rndr
@test minval(output) === 0.0
@test maxval(output) === 10.0
@test renderer(output).zerocolor == Image(zerocolor=(1.0, 0.0, 0.0)).zerocolor
@test isstored(output) == true
simdata = SimData(output, Ruleset(Life()))
# Test level normalisation
normed = normalise.(output[1][:a], minval(output), maxval(output))
@test normed == [0.8 1.0
0.0 0.5]
# Test greyscale Image conversion
img = render!(output, simdata)
@test img == [ARGB32(0.1, 0.1, 0.1, 1.0) ARGB32(1.0, 1.0, 1.0, 1.0)
ARGB32(1.0, 0.0, 0.0, 1.0) ARGB32(0.5, 0.5, 0.5, 1.0)]
output = NoDisplayImageOutput((a=init_,);
tspan=DateTime(2001):Year(1):DateTime(2010), mask=mask_,
renderer=Image(; scheme=leonardo), text=nothing,
minval=0.0, maxval=10.0, store=true
)
img = render!(output, simdata)
@test img == [DynamicGrids.MASKCOL l1
DynamicGrids.ZEROCOL l05]
z0 = ARGB32(1, 0, 0)
output = NoDisplayImageOutput((a=init_,);
tspan=DateTime(2001):Year(1):DateTime(2010), mask=mask_,
renderer = Image(scheme=leonardo, zerocolor=z0), text=nothing,
minval=0.0, maxval=10.0, store=true
)
img = render!(output, simdata)
@test img == [DynamicGrids.MASKCOL l1
z0 l05]
@testset "text captions" begin
pixelsize = 20
timepos = 2pixelsize, pixelsize
textinit = zeros(200, 200)
font = "arial"
face = findfont(font)
# Swap fonts on linux
if face === nothing
font = "sans-serif"
face = findfont(font)
end
if face !== nothing
refimg = ARGB32.(map(x -> ARGB32(1.0, 0.0, 0.0, 1.0), textinit))
renderstring!(refimg, string(DateTime(2001)), face, pixelsize, timepos...;
fcolor=ARGB32(1.0, 1.0, 1.0, 1.0), bcolor=ARGB32(0.0, 0.0, 0.0, 1.0))
textconfig=TextConfig(; font=font, timepixels=pixelsize, namepixels=pixelsize, bcolor=ARGB32(0))
output = NoDisplayImageOutput(textinit;
tspan=DateTime(2001):Year(1):DateTime(2001),
renderer=Image(zerocolor=ARGB32(1.0, 0.0, 0.0, 1.0)),
text=textconfig, store=true,
)
simdata = SimData(output, Ruleset())
img = render!(output, simdata);
@test img == refimg
end
end
end
@testset "SparseOptInspector" begin
init = Bool[
0 0 0 0 0 0 0
0 0 0 0 1 1 1
0 0 0 0 0 0 1
0 0 0 0 0 1 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
]
ruleset = Ruleset(Life();
timestep=Day(1),
boundary=Wrap(),
opt=SparseOpt(),
)
rndr = SparseOptInspector()
output = NoDisplayImageOutput(init;
tspan=Date(2001, 1, 1):Day(1):Date(2001, 1, 5),
renderer=rndr, minval=0.0, maxval=1.0, store=true
)
@test minval(output) === 0.0
@test maxval(output) === 1.0
@test renderer(output) == SparseOptInspector()
@test isstored(output) == true
global images = []
sim!(output, ruleset)
w, y, c = ARGB32(1), ARGB32(.0, .0, .5), ARGB32(.5, .5, .5)
@test_broken images[1] == [
y y y y y y y
y y y c w w w
y y y c c c w
y y y y y w c
y y y y y c c
y y y y y y y
]
end
@testset "Layout Renderer" begin
init = [8.0 10.0;
0.0 5.0]
z0 = DynamicGrids.ZEROCOL
grey = Greyscale()
multiinit = (a=init, b=2init)
rndr = Layout([:a nothing :b], [grey nothing leonardo])
@test DynamicGrids.imagesize(rndr, init, 1:1) == (2, 6)
output = NoDisplayImageOutput(multiinit;
tspan=DateTime(2001):Year(1):DateTime(2002),
renderer=rndr,
text=nothing,
minval=[0 nothing 0], maxval=[10 nothing 20],
store=true
)
@test minval(output) == [0 nothing 0]
@test maxval(output) == [10 nothing 20]
@test renderer(output) === rndr
@test isstored(output) == true
simdata = SimData(output, Ruleset(Life()))
# Test image is joined from :a, nothing, :b
@test render!(output, simdata) ==
[ARGB32(0.8, 0.8, 0.8, 1.0) ARGB32(1.0, 1.0, 1.0, 1.0) ARGB32(0.0) ARGB32(0.0) l08 l1
z0 ARGB32(0.5, 0.5, 0.5, 1.0) ARGB32(0.0) ARGB32(0.0) z0 l05]
@testset "text captions" begin
timepixels = 20
timepos = 2timepixels, timepixels
textinit = (a=zeros(200, 200), b=zeros(200, 200))
font = "arial"
face = findfont(font)
if face === nothing
font = "cantarell"
face = findfont(font)
end
if face !== nothing
# Set up refernce image
refimg = cat(fill(z0, 200, 200),
fill(ARGB32(0), 200, 200),
fill(z0, 200, 200); dims=1)
renderstring!(refimg, string(DateTime(2001)), face, timepixels, timepos...;
fcolor=ARGB32(RGB(1.0), 1.0), bcolor=ARGB32(RGB(0.0), 1.0))
namepixels = 15
nameposa = 3timepixels + namepixels, timepixels
renderstring!(refimg, "a", face, namepixels, nameposa...;
fcolor=ARGB32(RGB(1.0), 1.0), bcolor=ARGB32(RGB(0.0), 1.0))
nameposb = 3timepixels + namepixels + 400, timepixels
renderstring!(refimg, "b", face, namepixels, nameposb...;
fcolor=ARGB32(RGB(1.0), 1.0), bcolor=ARGB32(RGB(0.0), 1.0))
textconf = TextConfig(; font=font, timepixels=timepixels, namepixels=namepixels, bcolor=ARGB32(0))
# Build renderer
output = NoDisplayImageOutput(textinit;
tspan=DateTime(2001):Year(1):DateTime(2001),
text=textconf,
store=true,
layout=[:a, nothing, :b],
scheme=[grey, nothing, leonardo],
minval=[0, nothing, 0],
maxval=[1, nothing, 1]
)
output.imageconfig.renderer
simdata = SimData(output, Ruleset())
img = render!(output, simdata);
@test img == refimg
end
end
@testset "errors" begin
output = NoDisplayImageOutput(multiinit;
tspan=1:10, renderer=rndr,
minval=[0, 0, 0],
maxval=[10, 20],
)
simdata = SimData(output, Ruleset(Life()))
@test_throws ArgumentError render!(output, simdata)
broken_rndr = Layout([:d, :c], (grey, leonardo))
output = NoDisplayImageOutput(multiinit;
tspan=1:10, renderer=broken_rndr, text=nothing, minval=[0, 0], maxval=[10, 20],
)
simdata = SimData(output, Ruleset(Life()))
@test_throws ArgumentError render!(output, simdata)
end
@testset "Layout is the default for NamedTuple of grids" begin
output = NoDisplayImageOutput(multiinit; tspan=1:10)
@test renderer(output) isa Layout
@test DynamicGrids.imagesize(renderer(output), multiinit, 1:1) == (2, 4)
end
end
@testset "simulation savegif from ArrayOutput" begin
init_ = Bool[
0 0 0 0 0
0 0 1 1 1
0 0 0 0 1
0 0 0 1 0
]
ig = Image(zerocolor=RGB(0.0))
output = ArrayOutput(init_; tspan=1:10)
@test minval(output) == 0
@test maxval(output) == 1
@test renderer(output) isa Image
@test fps(output) === nothing
sim!(output, Life())
savegif("test2.gif", output; renderer=Image(zerocolor=RGB(0.0)), text=nothing)
gif = load("test2.gif")
@test gif[:, :, 1] == RGB.(cat(output...; dims=3))[:, :, 1]
rm("test2.gif")
savegif("test2.gif", output; zerocolor=RGB(0.0))
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 16497 | using DynamicGrids, DimensionalData, Test, Dates, Unitful,
CUDAKernels, FileIO, FixedPointNumbers, Colors
using DynamicGrids: Extent, SimData, gridview
if CUDAKernels.CUDA.has_cuda_gpu()
CUDAKernels.CUDA.allowscalar(false)
hardware = (SingleCPU(), ThreadedCPU(), CPUGPU(), CuGPU())
else
hardware = (SingleCPU(), ThreadedCPU(), CPUGPU())
end
opts = (NoOpt(), SparseOpt())
# proc = CPUGPU()
proc = SingleCPU()
# opt = SparseOpt()
opt = NoOpt()
# life glider sims
# Test all cycled variants of the array
cyclei!(arrays) = begin
for A in arrays
v = A[1, :]
@inbounds copyto!(A, CartesianIndices((1:size(A, 1)-1, 1:size(A, 2))),
A, CartesianIndices((2:size(A, 1), 1:size(A, 2))))
A[end, :] = v
end
end
cyclej!(arrays) = begin
for A in arrays
v = A[:, 1]
@inbounds copyto!(A, CartesianIndices((1:size(A, 1), 1:size(A, 2)-1)),
A, CartesianIndices((1:size(A, 1), 2:size(A, 2))))
A[:, end] = v
end
end
test6_7 = (
init = Bool[
0 0 0 0 0 0 0
0 0 0 0 1 1 1
0 0 0 0 0 0 1
0 0 0 0 0 1 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
],
test2 = Bool[
0 0 0 0 0 1 0
0 0 0 0 0 1 1
0 0 0 0 1 0 1
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
],
test3 = Bool[
0 0 0 0 0 1 1
0 0 0 0 1 0 1
0 0 0 0 0 0 1
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
],
test4 = Bool[
0 0 0 0 0 1 1
1 0 0 0 0 0 1
0 0 0 0 0 1 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
],
test5 = Bool[
1 0 0 0 0 1 1
1 0 0 0 0 0 0
0 0 0 0 0 0 1
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
],
test7 = Bool[
1 0 0 0 0 1 0
1 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
1 0 0 0 0 0 1
]
)
test5_6 = (
init = DimArray(Bool[
0 0 0 0 0 0
0 0 0 1 1 1
0 0 0 0 0 1
0 0 0 0 1 0
0 0 0 0 0 0
], (Y, X)),
test2 = Bool[
0 0 0 0 1 0
0 0 0 0 1 1
0 0 0 1 0 1
0 0 0 0 0 0
0 0 0 0 0 0
],
test3 = Bool[
0 0 0 0 1 1
0 0 0 1 0 1
0 0 0 0 0 1
0 0 0 0 0 0
0 0 0 0 0 0
],
test4 = Bool[
0 0 0 0 1 1
1 0 0 0 0 1
0 0 0 0 1 0
0 0 0 0 0 0
0 0 0 0 0 0
],
test5 = Bool[
1 0 0 0 1 1
1 0 0 0 0 0
0 0 0 0 0 1
0 0 0 0 0 0
0 0 0 0 0 0
],
test7 = Bool[
1 0 0 0 1 0
1 0 0 0 0 0
0 0 0 0 0 0
0 0 0 0 0 0
1 0 0 0 0 1
]
)
test = test5_6
@testset "Life simulation Wrap" begin
# Test on two sizes to test half blocks on both axes
# Loop over shifing init arrays to make sure they all work
for test in (test5_6, test6_7), i in 1:size(test[:init], 1)
for j in 1:size(test[:init], 2)
for proc in hardware, opt in opts
tspan = Date(2001, 1, 1):Day(2):Date(2001, 1, 14)
ruleset = Ruleset(;
rules=(Life(),),
timestep=Day(2),
boundary=Wrap(),
proc=proc,
opt=opt,
)
@testset "$(nameof(typeof(proc))) $(nameof(typeof(opt))) results match glider behaviour" begin
output = ArrayOutput(test[:init], tspan=tspan)
sim!(output, ruleset)
@test output[2] == test[:test2]
# || (println(2); display(output[2]); display(test[:test2]))
@test output[3] == test[:test3] # || (println(3); display(output[3]); display(test[:test3]))
@test output[4] == test[:test4] # || (println(4); display(output[4]); display(test[:test4]))
@test output[5] == test[:test5] # || (println(5); display(output[5]); display(test[:test5]))
@test output[7] == test[:test7] # || (println(7); display(output[7]); display(test[:test7]))
end
@testset "$(nameof(typeof(proc))) $(nameof(typeof(opt))) using step!" begin
simdata = DynamicGrids._proc_setup(SimData(Extent(; init=test[:init], tspan=tspan), ruleset))
# Need Array here to copy from GPU to CPU
@test Array(gridview(first(simdata))) == test[:init]
simdata = step!(simdata)
Array(gridview(first(simdata)))
@test Array(gridview(first(simdata))) == test[:test2] || (println("s2"); display(Array(gridview(first(simdata)))); display(test[:test2]))
simdata = step!(simdata)
@test Array(gridview(first(simdata))) == test[:test3] || (println("s3"); display(Array(gridview(first(simdata)))); display(test[:test3]))
simdata = step!(simdata)
@test Array(gridview(first(simdata))) == test[:test4] || (println("s4"); display(Array(gridview(first(simdata)))); display(test[:test4]))
simdata = step!(simdata)
@test Array(gridview(first(simdata))) == test[:test5] || (println("s5"); display(Array(gridview(first(simdata)))); display(test[:test5]))
simdata = step!(simdata)
simdata = step!(simdata)
@test Array(gridview(first(simdata))) == test[:test7] || (println("s7"); display(Array(gridview(first(simdata)))); display(test[:test7]))
end
end
cyclej!(test)
end
cyclei!(test)
end
nothing
end
@testset "Life simulation with Remove boudary" begin
init_ = DimArray(Bool[
0 0 0 0 0 0 0
0 0 0 0 1 1 1
0 0 0 0 0 0 1
0 0 0 0 0 1 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
], (X, Y))
test2_rem = Bool[
0 0 0 0 0 1 0
0 0 0 0 0 1 1
0 0 0 0 1 0 1
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
]
test3_rem = Bool[
0 0 0 0 0 1 1
0 0 0 0 1 0 1
0 0 0 0 0 0 1
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
]
test4_rem = Bool[
0 0 0 0 0 1 1
0 0 0 0 0 0 1
0 0 0 0 0 1 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
]
test5_rem = Bool[
0 0 0 0 0 1 1
0 0 0 0 0 0 1
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
]
test7_rem = Bool[
0 0 0 0 0 1 1
0 0 0 0 0 1 1
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
]
rule = Life{:a,:a}(neighborhood=Moore(1))
@testset "Wrong timestep throws an error" begin
rs = Ruleset(rule; timestep=Day(2), boundary=Remove(), opt=NoOpt())
output = ArrayOutput((a=init_,); tspan=1:7)
@test_throws ArgumentError sim!(output, rs; tspan=Date(2001, 1, 1):Month(1):Date(2001, 3, 1))
end
@testset "Results match glider behaviour" begin
output = ArrayOutput((a=init_,); tspan=1:7)
for proc in hardware, opt in opts
sim!(output, rule; boundary=Remove(), proc=proc, opt=opt)
output[2][:a]
output[3][:a]
output[4][:a]
output[5][:a]
output[7][:a]
@test output[2][:a] == test2_rem
@test output[3][:a] == test3_rem
@test output[4][:a] == test4_rem
@test output[5][:a] == test5_rem
@test output[7][:a] == test7_rem
end
end
@testset "Combinatoric comparisons in a larger Life sim" begin
rule = Life(neighborhood=Moore(1))
init_ = rand(Bool, 100, 99)
mask_ = ones(Bool, size(init_)...)
mask_[1:50, 1:50] .= false
wrap_rs_ref = Ruleset(rule; boundary=Wrap())
remove_rs_ref = Ruleset(rule; boundary=Remove())
wrap_output_ref = ArrayOutput(init_; tspan=1:100, mask=mask_)
remove_output_ref = ArrayOutput(init_; tspan=1:100, mask=mask_)
sim!(remove_output_ref, remove_rs_ref)
sim!(wrap_output_ref, wrap_rs_ref)
for proc in hardware, opt in opts
@testset "$(nameof(typeof(opt))) $(nameof(typeof(proc)))" begin
@testset "Wrap" begin
wrap_rs = Ruleset(rule; boundary=Wrap(), proc=proc, opt=opt)
wrap_output = ArrayOutput(init_; tspan=1:100, mask=mask_)
sim!(wrap_output, wrap_rs)
wrap_output_ref[2] .- wrap_output[2]
@test wrap_output_ref[2] == wrap_output[2]
wrap_output_ref[3] .- wrap_output[3]
@test wrap_output_ref[3] == wrap_output[3]
@test wrap_output_ref[10] == wrap_output[10]
@test wrap_output_ref[100] == wrap_output[100]
end
@testset "Remove" begin
remove_rs = Ruleset(rule; boundary=Remove(), proc=proc, opt=opt)
remove_output = ArrayOutput(init_; tspan=1:100, mask=mask_)
sim!(remove_output, remove_rs);
@test remove_output_ref[2] == remove_output[2]
@test remove_output_ref[3] == remove_output[3]
remove_output_ref[3] .- remove_output[3]
@test remove_output_ref[10] == remove_output[10]
@test remove_output_ref[100] == remove_output[100]
end
end
end
end
end
@testset "sim! with other outputs" begin
for proc in hardware, opt in opts
@testset "$(nameof(typeof(opt))) $(nameof(typeof(proc)))" begin
@testset "Transformed output" begin
ruleset = Ruleset(Life();
timestep=Month(1),
boundary=Wrap(),
proc=proc,
opt=opt,
)
tspan_ = Date(2010, 4):Month(1):Date(2010, 7)
output = TransformedOutput(sum, test6_7[:init]; tspan=tspan_)
sim!(output, ruleset)
@test output[1] == sum(test6_7[:init])
@test output[2] == sum(test6_7[:test2])
@test output[3] == sum(test6_7[:test3])
@test output[4] == sum(test6_7[:test4])
end
@testset "REPLOutput block works, in Unitful.jl seconds" begin
ruleset = Ruleset(;
rules=(Life(),),
timestep=5u"s",
boundary=Wrap(),
proc=proc,
opt=opt,
)
output = REPLOutput(test6_7[:init];
tspan=0u"s":5u"s":6u"s", style=Block(), fps=1000, store=true
)
@test DynamicGrids.isstored(output) == true
sim!(output, ruleset)
resume!(output, ruleset; tstop=30u"s")
@test output[At(5u"s")] == test6_7[:test2]
@test output[At(10u"s")] == test6_7[:test3]
@test output[At(20u"s")] == test6_7[:test5]
@test output[At(30u"s")] == test6_7[:test7]
end
@testset "REPLOutput braile works, in Months" begin
ruleset = Ruleset(Life();
timestep=Month(1),
boundary=Wrap(),
proc=proc,
opt=opt,
)
tspan_ = Date(2010, 4):Month(1):Date(2010, 7)
output = REPLOutput(test6_7[:init]; tspan=tspan_, style=Braile(), fps=1000, store=false)
sim!(output, ruleset)
@test output[At(Date(2010, 7))] == test6_7[:test4]
@test DynamicGrids.tspan(output) == Date(2010, 4):Month(1):Date(2010, 7)
resume!(output, ruleset; tstop=Date(2010, 10))
@test DynamicGrids.tspan(output) == Date(2010, 4):Month(1):Date(2010, 10)
@test output[1] == test6_7[:test7]
end
end
end
end
@testset "GifOutput saves" begin
@testset "Image generator" begin
# TODO fix on CUDA: cell_to_rgb indexes a CuArray
ruleset = Ruleset(;
rules=(Life(),),
boundary=Wrap(),
timestep=5u"s",
opt=NoOpt(),
)
output = GifOutput(test6_7[:init];
filename="test_gifoutput.gif", text=nothing,
tspan=0u"s":5u"s":30u"s", fps=10, store=true,
)
@test output.imageconfig.renderer isa Image
@test output.imageconfig.textconfig == nothing
@test DynamicGrids.isstored(output) == true
sim!(output, ruleset)
@test output[At(5u"s")] == test6_7[:test2]
@test output[At(10u"s")] == test6_7[:test3]
@test output[At(20u"s")] == test6_7[:test5]
@test output[At(30u"s")] == test6_7[:test7]
gif = load("test_gifoutput.gif")
@test gif == RGB.(output.gif)
rm("test_gifoutput.gif")
end
@testset "Layout" begin
# TODO fix on CUDA
zeroed = test6_7[:init]
ruleset = Ruleset(Life{:a}(); boundary=Wrap())
output = GifOutput((a=test6_7[:init], b=zeroed);
filename="test_gifoutput2.gif", text=nothing,
tspan=0u"s":5u"s":30u"s", fps=10, store=true
)
@test DynamicGrids.isstored(output) == true
@test output.imageconfig.renderer isa Layout
@test output.imageconfig.textconfig == nothing
sim!(output, ruleset)
@test all(map(==, output[At(5u"s")], (a=test6_7[:test2], b=zeroed)))
@test all(map(==, output[At(10u"s")], (a=test6_7[:test3], b=zeroed)))
@test all(map(==, output[At(20u"s")], (a=test6_7[:test5], b=zeroed)))
@test all(map(==, output[At(30u"s")], (a=test6_7[:test7], b=zeroed)))
gif = load("test_gifoutput2.gif")
@test gif == RGB.(output.gif)
@test gif[:, 1, 7] == RGB{N0f8}.([1.0, 1.0, 0.298, 0.298, 0.298, 1.0])
rm("test_gifoutput2.gif")
end
end
@testset "SparseOpt rules run everywhere with non zero values" begin
set_hood = SetNeighbors() do data, hood, val, I
for p in positions(hood, I)
data[p...] = 2
end
end
clearcell = Cell() do data, val, I
zero(val)
end
output = ArrayOutput(ones(10, 11); tspan=1:3)
sim!(output, set_hood; opt=SparseOpt())
@test all(output[3] .=== 2.0)
sim!(output, set_hood, clearcell; opt=SparseOpt())
@test all(output[3] .=== 0.0)
end
@testset "Single dimension rules" begin
init = zeros(Int, 7)
init[6] = true
rule110 = Neighbors(Moore(1; ndims=1)) do data, hood, c, I
l, r = neighbors(hood)
(c + r + c * r + l * c * r) % 2
end
REPLOutput(init; store=false, tspan=1:5, fps=100)
output = REPLOutput(init; tspan=1:5, fps=100)
@test DynamicGrids.isstored(output)
sim!(output, rule110)
tests = (
[0, 0, 0, 0, 0, 1, 0,],
[0, 0, 0, 0, 1, 1, 0,],
[0, 0, 0, 1, 1, 1, 0,],
[0, 0, 1, 1, 0, 1, 0,],
[0, 1, 1, 1, 1, 1, 0,],
)
map(tests, output) do t, o
@test t == o
end
init = zeros(Int, 50)
init[50] = true
filename = "test_1d_gifoutput.gif"
output = GifOutput(init; filename=filename, text=nothing, tspan=1:50)
sim!(output, rule110)
simgif = load(filename)
@test simgif == RGB.(output.gif)
rm(filename)
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 6967 | using DynamicGrids, Setfield, Test, LinearAlgebra, StaticArrays, OffsetArrays
using DynamicGrids.Neighborhoods
import DynamicGrids.Neighborhoods: _window, hoodsize, radius
init = [0 0 0 1 1 1
1 0 1 1 0 1
0 1 1 1 1 1
0 1 0 0 1 0
0 0 0 0 1 1
0 1 0 1 1 0]
win1 = [0 0 0
0 1 0
0 0 0]
win2 = [1 1 1
1 0 1
1 1 1]
win3 = [1 1 1
0 0 1
0 0 1]
@testset "Moore" begin
moore = Moore{1}(init[1:3, 1:3])
@test _window(moore) == init[1:3, 1:3]
@test hoodsize(moore) == 3
@test moore[1] == 0
@test length(moore) == 8
@test eltype(moore) == Int
@test neighbors(moore) isa Tuple
@test collect(neighbors(moore)) == [0, 1, 0, 0, 1, 0, 1, 1]
@test sum(moore) == sum(neighbors(moore)) == 4
@test offsets(moore) ==
((-1, -1), (0, -1), (1, -1), (-1, 0), (1, 0), (-1, 1), (0, 1), (1, 1))
moore1 = @set moore._window = win1
@test moore1._window == win1
moore2 = DynamicGrids.setwindow(moore, win2)
@test moore2._window == win2
@test sum(Moore{1}(win1)) == 0
@test sum(Moore{1}(win2)) == 8
@test sum(Moore{1}(win3)) == 5
end
@testset "Window" begin
@test Window{1}() == Window(1) == Window(zeros(3, 3))
window = Window{1}(init[1:3, 1:3])
@test _window(window) == init[1:3, 1:3]
@test hoodsize(window) == 3
@test window[1] == 0
@test window[2] == 1
@test length(window) == 9
@test eltype(window) == Int
@test neighbors(window) isa Array
@test neighbors(window) == _window(window)
@test sum(window) == sum(neighbors(window)) == 4
@test offsets(window) == ((-1, -1), (0, -1), (1, -1), (-1, 0), (0, 0),
(1, 0), (-1, 1), (0, 1), (1, 1))
window1 = @set window._window = win1
@test window1._window == win1
window2 = DynamicGrids.setwindow(window, win2)
@test window2._window == win2
@test sum(Window{1}(win1)) == 1
@test sum(Window{1}(win2)) == 8
@test sum(Window{1}(win3)) == 5
end
@testset "VonNeumann" begin
vonneumann = VonNeumann(1, init[1:3, 1:3])
@test offsets(vonneumann) == ((0, -1), (-1, 0), (1, 0), (0, 1))
@test _window(vonneumann) == init[1:3, 1:3]
@test hoodsize(vonneumann) == 3
@test vonneumann[1] == 1
@test vonneumann[2] == 0
@test length(vonneumann) == 4
@test eltype(vonneumann) == Int
@test neighbors(vonneumann) isa Tuple
@test collect(neighbors(vonneumann)) == [1, 0, 1, 1]
@test sum(neighbors(vonneumann)) == 3
vonneumann2 = VonNeumann(2)
@test offsets(vonneumann2) ==
((0, -2), (-1, -1), (0, -1), (1, -1),
(-2 , 0), (-1, 0), (1, 0), (2, 0),
(-1, 1), (0, 1), (1, 1), (0, 2))
vonneumann1 = @set vonneumann._window = win2
@test vonneumann1._window == win2
vonneumann2 = DynamicGrids.setwindow(vonneumann, win3)
@test vonneumann2._window == win3
@test sum(VonNeumann(1, win1)) == 0
@test sum(VonNeumann(1, win2)) == 4
@test sum(VonNeumann(1, win3)) == 2
end
@testset "Positional" begin
win = [0 1 0 0 1
0 0 1 0 0
0 0 0 1 1
0 0 1 0 1
1 0 1 0 1]
custom1 = Positional(((-1,-1), (2,-2), (2,2), (-1,2), (0,0)), win)
custom2 = Positional{((-1,-1), (0,-1), (1,-1), (2,-1), (0,0))}(win)
layered = LayeredPositional(
(Positional((-1,1), (-2,2)), Positional((1,2), (2,2))), win)
@test neighbors(custom1) isa Tuple
@test collect(neighbors(custom1)) == [0, 1, 1, 0, 0]
@test sum(custom1) == 2
@test sum(custom2) == 0
@test sum(layered) == (1, 2)
@test offsets(layered) == (((-1, 1), (-2, 2)), ((1, 2), (2, 2)))
layered1 = @set layered._window = 2win
@test layered1._window == 2win
layered2 = DynamicGrids.setwindow(layered, 3win)
@test layered2._window == 3win
win = reshape(2:10, 3, 3)
hood = Positional(((-1, -1), (1, 1)), win)
@test neighbors(hood) == (2, 10)
@test offsets(hood) == ((-1, -1), (1, 1))
@test positions(hood, (2, 2)) == ((1, 1), (3, 3))
end
@testset "LayeredPositional" begin
lhood = LayeredPositional(
Positional(((-1, -1), (1, 1)), ), Positional(((-2, -2), (2, 2)), )
)
@test offsets(lhood) == (((-1, -1), (1, 1)), ((-2, -2), (2, 2)))
@test collect.(collect(positions(lhood, (1, 1)))) ==
[[(0, 0), (2, 2)],
[(-1, -1), (3, 3)]]
win = reshape(1:25, 5, 5)
lhood_win = DynamicGrids.setwindow(lhood, win)
@test lhood_win._window == lhood_win.layers[1]._window ===
lhood_win.layers[2]._window === win
lhood_win.layers[2]._window
@test map(radius, lhood_win.layers) == (2, 2)
@test neighbors(lhood_win) == ((7, 19), (1, 25))
end
@testset "Kernel" begin
win = reshape(1:9, 3, 3)
@testset "Window" begin
mat = zeros(3, 3)
@test Kernel(mat) == Kernel(Window(1), mat)
@test_throws ArgumentError Kernel(Window(2), mat)
k = Kernel(Window{1,2,9,typeof(win)}(win), SMatrix{3,3}(reshape(1:9, 3, 3)))
@test kernelproduct(k) == sum((1:9).^2)
@test neighbors(k) == reshape(1:9, 3, 3)
@test offsets(k) == ((-1, -1), (0, -1), (1, -1), (-1, 0), (0, 0),
(1, 0), (-1, 1), (0, 1), (1, 1))
@test positions(k, (2, 2)) == ((1, 1), (2, 1), (3, 1), (1, 2),
(2, 2), (3, 2), (1, 3), (2, 3), (3, 3))
end
@testset "Moore" begin
k = Kernel(Moore{1,2,8,typeof(win)}(win), (1:4..., 6:9...))
@test kernelproduct(k) == sum((1:4).^2) + sum((6:9).^2)
end
@testset "Positional" begin
off = ((0,-1),(-1,0),(1,0),(0,1))
hood = Positional{off,1,2,4,typeof(win)}(win)
k = Kernel(hood, 1:4)
@test kernelproduct(k) == 1 * 2 + 2 * 4 + 3 * 6 + 4 * 8
end
end
@testset "neighbors works on rule" begin
rule = Life(;neighborhood=Moore{1}([0 1 1; 0 0 0; 1 1 1]))
@test sum(neighbors(rule)) == 5
end
@testset "readwindow" begin
grid1 = [0, 1, 2, 3, 4, 0]
grid2 = [
0 0 0 0 0 0
0 1 2 3 4 0
0 5 6 7 8 0
0 9 10 11 12 0
0 0 0 0 0 0
]
@test DynamicGrids.readwindow(Moore{1,1}(), grid1, (2,)) == [0, 1, 2]
@test DynamicGrids.readwindow(Moore{1,1}(), OffsetArray(grid1, (0:5)), (2,)) == [1, 2, 3]
@test_throws DimensionMismatch DynamicGrids.readwindow(Moore{1,1}(), grid2, (2,))
@test_throws DimensionMismatch DynamicGrids.readwindow(Moore{1,2}(), grid2, (2,))
@test DynamicGrids.readwindow(Moore{1,2}(), grid2, (2, 2)) == [0 0 0; 0 1 2; 0 5 6]
@test DynamicGrids.readwindow(Moore{1,2}(), OffsetArray(grid2, (0:4, 0:5)), (2, 2)) == [1 2 3; 5 6 7; 9 10 11]
end
@testset "pad/unpad axes" begin
A = zeros(6, 7)
@test pad_axes(A, 2) == (-1:8, -1:9)
@test pad_axes(A, Moore(3)) == (-2:9, -2:10)
@test unpad_axes(A, 2) == (3:4, 3:5)
@test unpad_axes(A, VonNeumann(1)) == (2:5, 2:6)
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 8914 | using DynamicGrids, StaticArrays, Test, FileIO, Colors, FixedPointNumbers
using DynamicGrids: SimData, NoDisplayImageOutput
@testset "CellRule that multiples a StaticArray" begin
rule = Cell{:grid1}() do data, state, I
2state
end
init = (grid1 = fill(SA[1.0, 2.0], 5, 5),)
output = ArrayOutput(init; tspan=1:3)
sim!(output, rule)
@test output[2][:grid1] == reshape([ # Have to use reshape to construct this
SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0],
SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0],
SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0],
SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0],
SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0], SA[2.0, 4.0]
], (5, 5))
@test output[3][:grid1] == reshape([ # Have to use reshape to construct this
SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0],
SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0],
SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0],
SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0],
SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0], SA[4.0, 8.0]
], (5, 5))
end
@testset "Neighborhood Rule that sums a Neighborhood of StaticArrays" begin
rule = Neighbors{Tuple{:grid1,:grid2},:grid1}(Moore(1)) do data, hood, (s1, s2), I
sum(hood) .+ s2
end
init = (
grid1 = fill(SA[1.0, 2.0], 5, 5),
grid2 = fill(0.5, 5, 5),
)
output = ArrayOutput(init; tspan=1:2)
sim!(output, rule)
@test output[2][:grid1] == reshape([ # Have to use reshape to construct this
SA[3.5, 6.5], SA[5.5, 10.5], SA[5.5, 10.5], SA[5.5, 10.5], SA[3.5, 6.5],
SA[5.5, 10.5], SA[8.5, 16.5], SA[8.5, 16.5], SA[8.5, 16.5], SA[5.5, 10.5],
SA[5.5, 10.5], SA[8.5, 16.5], SA[8.5, 16.5], SA[8.5, 16.5], SA[5.5, 10.5],
SA[5.5, 10.5], SA[8.5, 16.5], SA[8.5, 16.5], SA[8.5, 16.5], SA[5.5, 10.5],
SA[3.5, 6.5], SA[5.5, 10.5], SA[5.5, 10.5], SA[5.5, 10.5], SA[3.5, 6.5]
], (5, 5))
end
@testset "SetCell randomly updates a StaticArray" begin
rule = SetCell{:grid1}() do data, state, I
if I == (2, 2) || I == (1, 3)
data[:grid1][I...] = SA[99.0, 100.0]
end
end
init = (grid1 = fill(SA[0.0, 0.0], 3, 3),)
output = ArrayOutput(init; tspan=1:2)
sim!(output, rule)
@test output[2][:grid1] == reshape([ # Have to use reshape to construct this
SA[0.0, 0.0], SA[0.0, 0.0], SA[0.0, 0.0],
SA[0.0, 0.0], SA[99.0, 100.0], SA[0.0, 0.0],
SA[99.0, 100.0], SA[0.0, 0.0], SA[0.0, 0.0]
], (3, 3))
end
struct TestStruct{A,B}
a::A
b::B
end
const TS = TestStruct
Base.:*(ts::TestStruct, x::Number) = TestStruct(ts.a * x, ts.b * x)
Base.:*(x::Number, ts::TestStruct) = TestStruct(x * ts.a, x * ts.b)
Base.:/(ts::TestStruct, x::Number) = TestStruct(ts.a / x, ts.b / x)
Base.:+(ts1::TestStruct, ts2::TestStruct) = TestStruct(ts1.a + ts2.a, ts1.b + ts2.b)
Base.:-(ts1::TestStruct, ts2::TestStruct) = TestStruct(ts1.a - ts2.a, ts1.b - ts2.b)
Base.:+(ts::TestStruct, x::Number) = TestStruct(ts.a + x, ts.b + x)
Base.:-(ts::TestStruct, x::Number) = TestStruct(ts.a - x, ts.b - x)
Base.isless(a::TestStruct, b::TestStruct) = isless(a.a, b.a)
Base.zero(::Type{<:TestStruct{T1,T2}}) where {T1,T2} = TestStruct(zero(T1), zero(T2))
Base.oneunit(::Type{<:TestStruct{T1,T2}}) where {T1,T2} = TestStruct(oneunit(T1), oneunit(T2))
DynamicGrids.to_rgb(scheme::ObjectScheme, obj::TestStruct) = ARGB32(obj.a)
DynamicGrids.to_rgb(scheme, obj::TestStruct) = get(scheme, obj.a)
@testset "CellRule that multiples a struct" begin
rule = Cell{:grid1,:grid1}() do data, state, I
2state
end
init = (grid1 = fill(TS(1.0, 2.0), 5, 5),)
output = ArrayOutput(init; tspan=1:3)
sim!(output, rule)
@test output[2][:grid1] == reshape([ # Have to use reshape to construct this
TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0),
TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0),
TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0),
TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0),
TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0), TS(2.0, 4.0)
], (5, 5))
@test output[3][:grid1] == reshape([ # Have to use reshape to construct this
TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0),
TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0),
TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0),
TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0),
TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0)
], (5, 5))
end
@testset "Neighborhood Rule that sums a Neighborhood of stucts" begin
rule = Neighbors{Tuple{:grid1,:grid2},:grid1}(Moore(1)) do data, hood, (s1, s2), I
sum(hood) * s2
end
init = (
grid1 = fill(TS(1.0, 2.0), 5, 5),
grid2 = fill(0.5, 5, 5),
)
output = ArrayOutput(init; tspan=1:2)
sim!(output, rule)
@test output[2][:grid1] == reshape([ # Have to use reshape to construct this
TS(1.5, 3.0), TS(2.5, 5.0), TS(2.5, 5.0), TS(2.5, 5.0), TS(1.5, 3.0),
TS(2.5, 5.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(2.5, 5.0),
TS(2.5, 5.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(2.5, 5.0),
TS(2.5, 5.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(4.0, 8.0), TS(2.5, 5.0),
TS(1.5, 3.0), TS(2.5, 5.0), TS(2.5, 5.0), TS(2.5, 5.0), TS(1.5, 3.0),
], (5, 5))
end
@testset "SetCell rule randomly updates a struct" begin
rule = SetCell{:grid1,:grid1}() do data, state, I
if I == (2, 2) || I == (1, 3)
data[:grid1][I...] = TS(99.0, 100.0)
end
end
init = (grid1 = fill(TS(0.0, 0.0), 3, 3),)
output = ArrayOutput(init; tspan=1:2)
sim!(output, rule)
@test output[2][:grid1] == reshape([ # Have to use reshape to construct this
TS(0.0, 0.0), TS(0.0, 0.0), TS(0.0, 0.0),
TS(0.0, 0.0), TS(99.0, 100.0), TS(0.0, 0.0),
TS(99.0, 100.0), TS(0.0, 0.0), TS(0.0, 0.0)
], (3, 3))
end
@testset "object grid can generate an image" begin
@testset "normalise" begin
@test DynamicGrids.to_rgb(ObjectScheme(), TestStruct(99.0, 1.0) / 99) == ARGB32(1.0)
@test DynamicGrids.to_rgb(ObjectScheme(), TestStruct(00.0, 0.0) / 99) == ARGB32(0.0)
@test DynamicGrids.to_rgb(ObjectScheme(), DynamicGrids.normalise(TestStruct(99.0, 1.0), nothing, 99)) == ARGB32(1.0)
end
rule = SetCell{:grid1,:grid1}() do data, state, I
if I == (2, 2) || I == (1, 3)
data[:grid1][I...] = TS(99.0, 100.0)
end
end
init = (grid1=fill(TS(0.0, 0.0), 3, 3),)
# These should have the same answer
output1 = GifOutput(init;
filename="objectgrid.gif", store=true, tspan=1:2, maxval=[99.0], text=nothing
)
output2 = GifOutput(init;
filename="objectgrid_greyscale.gif", scheme=Greyscale(), store=true, tspan=1:2,
maxval=reshape([99.0], 1, 1), text=nothing
)
sim!(output1, rule)
sim!(output2, rule)
@test output1[2][:grid1] ==
[TS(0.0, 0.0) TS(0.0, 0.0) TS(99.0, 100.0)
TS(0.0, 0.0) TS(99.0, 100.0) TS(0.0, 0.0)
TS(0.0, 0.0) TS(0.0, 0.0) TS(0.0, 0.0)]
@test RGB.(output1.gif[:, :, 2]) ==
RGB.(output2.gif[:, :, 2]) ==
load("objectgrid.gif")[:, :, 2] ==
load("objectgrid_greyscale.gif")[:, :, 2] ==
map(xs -> RGB{N0f8}(xs...),
[(0.298,0.298,0.298) (0.298,0.298,0.298) (1.0,1.0,1.0)
(0.298,0.298,0.298) (1.0,1.0,1.0) (0.298,0.298,0.298)
(0.298,0.298,0.298) (0.298,0.298,0.298) (0.298,0.298,0.298)]
)
end
@testset "static arrays grid can generate an image" begin
rule = Cell{:grid1}() do data, state, I
2state
end
init = (grid1 = fill(SA[1.0, 2.0], 5, 5),)
# We can index into the SArray or access
# it with a function, defined using a Pair
output = GifOutput(init;
filename="sa.gif",
tspan=1:3,
store=true,
layout=[:grid1=>1 :grid1=>x->x[2]],
scheme=[Greyscale() Greyscale()],
minval=[0.0 0.0], maxval=[10.0 10.0],
text=nothing,
)
sim!(output, rule)
a02 = ARGB32(0.2)
a04 = ARGB32(0.4)
@test output.gif[:, :, 2] ==
[a02 a02 a02 a02 a02 a04 a04 a04 a04 a04
a02 a02 a02 a02 a02 a04 a04 a04 a04 a04
a02 a02 a02 a02 a02 a04 a04 a04 a04 a04
a02 a02 a02 a02 a02 a04 a04 a04 a04 a04
a02 a02 a02 a02 a02 a04 a04 a04 a04 a04]
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 1500 | using DynamicGrids, DimensionalData, Dates, Test
using DynamicGrids: isshowable, frameindex, storeframe!, SimData, stoppedframe
using DimensionalData.LookupArrays, DimensionalData.Dimensions
# Mostly outputs are tested in integration.jl
@testset "Output construction" begin
init = [10.0 11.0
0.0 5.0]
output = ArrayOutput(init; tspan=Date(2001):Year(1):Date(2010))
ruleset = Ruleset(Life())
@test length(output) == 10
@test size(output) == (10,)
@test step(output) == Year(1)
@test stoppedframe(output) == 10
@test timestep(output) == Year(1)
@test_throws ArgumentError DynamicGrids.ruleset(output)
@test frameindex(output, 5) == 5
@test isshowable(output, 5) == false
@test output[1] == output[Ti(1)] == init
@testset "DimensionalData interface" begin
@test output isa AbstractDimArray{<:Array,1,<:Tuple{<:Ti}}
@test dims(output) isa Tuple{<:Ti}
@test DimensionalData.name(output) == Symbol("")
@test metadata(output) == NoMetadata()
da = output[Ti(Between(Date(2002), Date(2003)))]
@test da isa DimArray{<:Array,1,<:Tuple{<:Ti}}
@test lookup(da) == (Date(2002):Year(1):Date(2002),)
end
@testset "errors" begin
@test_throws UndefKeywordError ArrayOutput(ones(5, 5))
@test_throws ArgumentError ArrayOutput((a=ones(5, 5), b=ones(4, 4)); tspan=1:10)
@test_throws ArgumentError ArrayOutput(ones(5, 5); mask=ones(2, 2), tspan=1:10)
end
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 9147 | using DynamicGrids, Dates, DimensionalData, Setfield, Unitful, Test
using Unitful: d
using DynamicGrids: SimData, Extent, _calc_auxframe, _cyclic_index
const DG = DynamicGrids
@testset "Aux" begin
@testset "sequence cycling" begin
@test _cyclic_index(-4, 2) == 2
@test _cyclic_index(-3, 2) == 1
@test _cyclic_index(-2, 2) == 2
@test _cyclic_index(-1, 2) == 1
@test _cyclic_index(0, 2) == 2
@test _cyclic_index(1, 2) == 1
@test _cyclic_index(2, 2) == 2
@test _cyclic_index(3, 2) == 1
@test _cyclic_index(4, 2) == 2
@test _cyclic_index(20, 10) == 10
@test _cyclic_index(21, 10) == 1
@test _cyclic_index(27, 10) == 7
end
@testset "aux sequence" begin
a = cat([0.1 0.2; 0.3 0.4], [1.1 1.2; 1.3 1.4], [2.1 2.2; 2.3 2.4]; dims=3)
@testset "the correct frame is calculated for aux data" begin
dimz = X(1:2), Y(1:2), Ti(15d:5d:25d)
seq = DimArray(a, dimz)
init = zero(seq[Ti(1)])
sd = SimData(Extent(init=init, aux=(seq=seq,), tspan=1d:1d:100d), Ruleset())
@test DynamicGrids.boundscheck_aux(sd, Aux{:seq}()) == true
tests = (1, 1), (4, 1), (5, 2), (6, 2), (9, 2), (10, 3), (11, 3), (14, 3), (15, 1),
(19, 1), (20, 2), (25, 3), (29, 3), (30, 1), (34, 1), (35, 2)
for (f, ref_af) in tests
@set! sd.currentframe = f
af = _calc_auxframe(sd).seq
@test af == ref_af
end
end
@testset "boundscheck_aux" begin
seq1 = zeros(Ti(15d:5d:25d))
seq3 = zeros((X(1:2), Y(1:2), Ti(15d:5d:25d)))
auxarray1 = zeros(X(3))
auxarray2 = zeros(dims(seq3, (X, Y)))
bigseq = zeros((X(1:5), Y(1:2), Ti(15d:5d:25d)))
aux1 = (seq=seq1, a1=auxarray1, a2=auxarray2)
sd1 = SimData(Extent(init=zeros(3), aux=aux1, tspan=1d:1d:100d), Ruleset())
aux2 = (seq=seq3, bigseq=bigseq, a1=auxarray1, a2=auxarray2)
sd2 = SimData(Extent(init=zero(seq3[Ti(1)]), aux=aux2, tspan=1d:1d:100d), Ruleset())
@test DynamicGrids.boundscheck_aux(sd1, Aux{:seq}()) == true
@test DynamicGrids.boundscheck_aux(sd1, Aux{:a1}()) == true
@test DynamicGrids.boundscheck_aux(sd2, Aux{:seq}()) == true
@test DynamicGrids.boundscheck_aux(sd2, Aux{:a2}()) == true
@test_throws ErrorException DynamicGrids.boundscheck_aux(sd1, Aux{:a2}())
@test_throws ErrorException DynamicGrids.boundscheck_aux(sd2, Aux{:a1}())
@test_throws ErrorException DynamicGrids.boundscheck_aux(sd2, Aux{:bigseq}())
@test_throws ErrorException DynamicGrids.boundscheck_aux(sd2, Aux{:missingseq}())
end
@testset "correct values are returned by get" begin
x, y, ti = X(1:2), Y(1:2), Ti(Date(2001, 1, 15):Day(5):Date(2001, 1, 25))
@testset "1d" begin
seq1 = DimArray(a[2, 2, :], ti)
seq3 = DimArray(a[:, 1, :], (x, ti))
init = zero(seq3[Ti(1)])
tspan = Date(2001):Day(1):Date(2001, 3)
data = SimData(Extent(init=init, aux=(; seq1, seq3), tspan=tspan), Ruleset())
data1 = DG._updatetime(data, 1)
@test data1.auxframe == (seq1 = 1, seq3 = 1,)
# I is ignored for 1d with Ti dim
@test get(data1, Aux(:seq1), (-10,)) == 0.4
@test get(data1, Aux(:seq3), (1)) == 0.1
dims(seq1, Ti) === dims(seq3, Ti)
data2 = DG._updatetime(data, 5);
@test data2.auxframe == (seq1 = 2, seq3 = 2,)
@test get(data2, Aux(:seq1), 11) == 1.4
@test get(data2, Aux(:seq3), 1) == 1.1
data3 = DG._updatetime(data, 10)
@test data3.auxframe == (seq1 = 3, seq3 = 3,)
@test get(data3, Aux(:seq1), (1,)) == 2.4
@test get(data3, Aux(:seq3), (1,)) == 2.1
data4 = DG._updatetime(data, 15)
@test data4.auxframe == (seq1 = 1, seq3 = 1,)
@test get(data4, Aux(:seq1), CartesianIndex(1,)) == 0.4
@test get(data4, Aux(:seq3), CartesianIndex(1,)) == 0.1
end
@testset "2d" begin
seq1 = DimArray(a[2, 2, :], ti)
seq3 = DimArray(a, (x, y, ti))
init = zero(seq3[Ti(1)])
tspan = Date(2001):Day(1):Date(2001, 3)
data = SimData(Extent(init=init, aux=(; seq1, seq3), tspan=tspan), Ruleset())
data1 = DG._updatetime(data, 1)
@test data1.auxframe == (seq1 = 1, seq3 = 1,)
# I is ignored for 1d with Ti dim
@test get(data1, Aux(:seq1), (-10, 17)) == 0.4
@test get(data1, Aux(:seq3), (1, 1)) == 0.1
dims(seq1, Ti) === dims(seq3, Ti)
data2 = DG._updatetime(data, 5);
@test data2.auxframe == (seq1 = 2, seq3 = 2,)
@test get(data2, Aux(:seq1), 11, 1) == 1.4
@test get(data2, Aux(:seq3), 1, 1) == 1.1
data3 = DG._updatetime(data, 10)
@test data3.auxframe == (seq1 = 3, seq3 = 3,)
@test get(data3, Aux(:seq1), (1, 10)) == 2.4
@test get(data3, Aux(:seq3), (1, 1)) == 2.1
data4 = DG._updatetime(data, 15)
@test data4.auxframe == (seq1 = 1, seq3 = 1,)
@test get(data4, Aux(:seq1), CartesianIndex(1, 1)) == 0.4
@test get(data4, Aux(:seq3), CartesianIndex(1, 1)) == 0.1
end
end
@testset "errors" begin
output = ArrayOutput(zeros(3, 3); tspan=1:3)
@test_throws ArgumentError DynamicGrids.aux(output, Aux{:somekey}())
end
end
end
# Use copyto to test all parametersources, as well as testing CopyTo itself
@testset "CopyTo" begin
init = [0 0]
@testset "Copy construction" begin
rule = CopyTo(7)
rule2 = @set rule.from = Aux{:a}()
@test rule2.from == Aux{:a}()
end
@testset "CopyTo from value" begin
ruleset = Ruleset(CopyTo(7))
output = ArrayOutput(init; tspan=1d:1d:3d)
sim!(output, ruleset)
@test output == [[0 0], [7 7], [7 7]]
end
@testset "CopyTo from Grid" begin
@test CopyTo(Aux(:l)) === CopyTo(; from=Aux(:l))
@test CopyTo{:a}(; from=Aux(:l)) === CopyTo{:a}(Aux(:l))
ruleset = Ruleset(CopyTo(Aux(:l)))
output = ArrayOutput(init; tspan=1:3, aux=(l=[3 4],))
sim!(output, ruleset)
@test output == [[0 0], [3 4], [3 4]]
da = DimArray(cat([1 2], [3 4]; dims=3) , (X(), Y(), Ti(4d:1d:5d)))
output = ArrayOutput(init; tspan=1d:1d:3d, aux=(l=da,))
sim!(output, ruleset)
@test output == [[0 0], [1 2], [3 4]]
end
@testset "CopyTo from Grid" begin
ruleset = Ruleset(Cell{:s,:s}((d, x, I) -> x + 1), CopyTo{:d}(from=Grid(:s)))
output = ArrayOutput((s=[1 3], d=[0 0],); tspan=1d:1d:3d)
sim!(output, ruleset)
@test output == [(s=[1 3], d=[0 0]), (s=[2 4], d=[2 4]), (s=[3 5], d=[3 5])]
ruleset = Ruleset(Cell{:s,:s}((d, x, I) -> x + 1), CopyTo{Tuple{:d1,:d2}}(from=Grid{:s}()))
output = ArrayOutput((s=[1 3], d1=[0 0], d2=[-1 -1],); tspan=1d:1d:3d)
sim!(output, ruleset)
@test output == [(s=[1 3], d1=[0 0], d2=[-1 -1]),
(s=[2 4], d1=[2 4], d2=[2 4]),
(s=[3 5], d1=[3 5], d2=[3 5])]
end
@testset "CopyTo from Delay" begin
ruleset = Ruleset(Cell{:s,:s}((d, x, I) -> x + 1), CopyTo{:d}(from=Delay{:s}(1d)))
@test DynamicGrids.hasdelay(rules(ruleset)) == true
output = ArrayOutput((s=[1 3], d=[0 0],); tspan=1d:1d:4d)
sim!(output, ruleset)
@test output == [
(s=[1 3], d=[0 0]),
(s=[2 4], d=[1 3]),
(s=[3 5], d=[2 4]),
(s=[4 6], d=[3 5])
]
ruleset = Ruleset(Cell{:s,:s}((d, x, I) -> x + 1), CopyTo{:d}(from=Delay{:s}(Month(2))))
@test DynamicGrids.hasdelay(rules(ruleset)) == true
output = ArrayOutput((s=[1 3], d=[0 0]); tspan=Date(2001):Month(1):Date(2001, 6))
sim!(output, ruleset)
@test output == [
(s=[1 3], d=[0 0]),
(s=[2 4], d=[1 3]),
(s=[3 5], d=[1 3]),
(s=[4 6], d=[2 4]),
(s=[5 7], d=[3 5]),
(s=[6 8], d=[4 6]),
]
end
@testset "CopyTo from Lag" begin
ruleset = Ruleset(Cell{:s,:s}((d, x, I) -> x + 1), CopyTo{:d}(from=Lag{:s}(1)))
@test DynamicGrids.hasdelay(rules(ruleset)) == true
output = ArrayOutput((s=[1 3], d=[0 0],); tspan=1d:1d:4d)
sim!(output, ruleset)
@test output == [
(s=[1 3], d=[0 0]),
(s=[2 4], d=[1 3]),
(s=[3 5], d=[2 4]),
(s=[4 6], d=[3 5])
]
end
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 19074 | using DynamicGrids, ModelParameters, Setfield, Test, StaticArrays,
LinearAlgebra, CUDAKernels
import DynamicGrids: applyrule, applyrule!, maprule!, ruletype, extent, source, dest,
_getreadgrids, _getwritegrids, _combinegrids, _readkeys, _writekeys,
SimData, WritableGridData, Rule, Extent, CPUGPU, neighborhoodkey
if CUDAKernels.CUDA.has_cuda_gpu()
CUDAKernels.CUDA.allowscalar(false)
hardware = (SingleCPU(), ThreadedCPU(), CPUGPU(), CuGPU())
else
hardware = (SingleCPU(), ThreadedCPU(), CPUGPU())
end
init = [0 1 1 0
0 1 1 0
0 1 1 0
0 1 1 0
0 1 1 0]
proc = SingleCPU()
opt = NoOpt()
@testset "Generic rule constructors" begin
rule1 = Cell{:a}(identity)
@test rule1.f == identity
@test_throws ArgumentError Cell()
rule2 = Cell{:a,:a}(identity)
@test rule1 == rule2
#@test_throws ArgumentError Cell(identity, identity)
rule1 = Neighbors{:a,:b}(identity, Moore(1))
@test rule1.f == identity
rule2 = Neighbors{:a,:b}(identity; neighborhood=Moore(1))
@test rule1 == rule2
@test typeof(rule1) == Neighbors{:a,:b,typeof(identity),Moore{1,2,8,Nothing}}
rule1 = Neighbors(identity, Moore(1))
@test rule1.f == identity
rule2 = Neighbors(identity; neighborhood=Moore(1))
@test typeof(rule1) == Neighbors{:_default_,:_default_,typeof(identity),Moore{1,2,8,Nothing}}
@test rule1 == rule2
@test_throws ArgumentError Neighbors()
# @test_throws ArgumentError Neighbors(identity, identity, identity)
rule1 = SetNeighbors{:a,:b}(identity, Moore(1))
@test rule1.f == identity
rule2 = SetNeighbors{:a,:b}(identity; neighborhood=Moore(1))
@test rule1 == rule2
@test typeof(rule1) == SetNeighbors{:a,:b,typeof(identity),Moore{1,2,8,Nothing}}
rule1 = SetNeighbors(identity, Moore(1))
@test rule1.f == identity
rule2 = SetNeighbors(identity; neighborhood=Moore(1))
@test typeof(rule1) == SetNeighbors{:_default_,:_default_,typeof(identity),Moore{1,2,8,Nothing}}
@test rule1 == rule2
@test_throws ArgumentError Neighbors()
# @test_throws ArgumentError Neighbors(identity, identity, identity)
rule1 = SetCell{:a,:b}(identity)
@test rule1.f == identity
@test_throws ArgumentError SetCell()
# @test_throws ArgumentError SetCell(identity, identity)
end
@testset "Rulesets" begin
rule1 = Cell(x -> 2x)
rule2 = Cell(x -> 3x)
rs1 = Ruleset((rule1, rule2); opt=NoOpt())
rs2 = Ruleset(rule1, rule2; opt=NoOpt())
@test rules(rs1) == rules(rs2)
@test typeof(Ruleset(StaticRuleset(rs1))) == typeof(rs1)
for fn in fieldnames(typeof(rs1))
@test getfield(Ruleset(StaticRuleset(rs1)), fn) == getfield(rs1, fn)
end
@test typeof(Ruleset(StaticRuleset(rs1))) == typeof(rs1)
ModelParameters.setparent!(rs2, (rule1,))
@test rs2.rules == (rule1,)
end
@testset "Cell" begin
rule = Cell((d, x, I) -> 2x)
@test applyrule(nothing, rule, 1, (0, 0)) == 2
end
@testset "Neighbors" begin
window = [1 0 0; 0 0 1; 0 0 1]
rule = Neighbors(VonNeumann(1, window)) do data, hood, state, I
sum(hood)
end
@test applyrule(nothing, rule, 0, (3, 3)) == 1
rule = Neighbors(Moore{1}(window)) do data, hood, state, I
sum(hood)
end
@test applyrule(nothing, rule, 0, (3, 3)) == 3
end
struct TestNeighborhoodRule{R,W,N} <: NeighborhoodRule{R,W}
neighborhood::N
end
DynamicGrids.applyrule(data, rule::TestNeighborhoodRule, state, index) = state
struct TestSetNeighborhoodRule{R,W,N} <: SetNeighborhoodRule{R,W}
neighborhood::N
end
function DynamicGrids.applyrule!(
data, rule::TestSetNeighborhoodRule{R,Tuple{W1,}}, state, index
) where {R,W1}
add!(data[W1], state[1], index...)
end
win5x5 = zeros(5, 5)
win7x7 = zeros(7, 7)
@testset "neighborhood rules" begin
ruleA = TestSetNeighborhoodRule{:a,:a}(Moore{3}(win7x7))
ruleB = TestSetNeighborhoodRule{Tuple{:b},Tuple{:b}}(Moore{2}(win5x5))
@test offsets(ruleA) isa Tuple
@test positions(ruleA, (1, 1)) isa Tuple
@test neighborhood(ruleA) == Moore{3}(win7x7)
@test neighborhood(ruleB) == Moore{2}(win5x5)
@test neighborhoodkey(ruleA) == :a
@test neighborhoodkey(ruleB) == :b
ruleA = TestNeighborhoodRule{:a,:a}(Moore{3}(win7x7))
ruleB = TestNeighborhoodRule{Tuple{:b},Tuple{:b}}(Moore{2}(win5x5))
@test offsets(ruleA) isa Tuple
@test neighborhood(ruleA) == Moore{3}(win7x7)
@test neighborhood(ruleB) == Moore{2}(win5x5)
@test neighborhoodkey(ruleA) == :a
@test neighborhoodkey(ruleB) == :b
@test offsets(ruleB) ===
((-2,-2), (-1,-2), (0,-2), (1,-2), (2,-2),
(-2,-1), (-1,-1), (0,-1), (1,-1), (2,-1),
(-2,0), (-1,0), (1,0), (2,0),
(-2,1), (-1,1), (0,1), (1,1), (2,1),
(-2,2), (-1,2), (0,2), (1,2), (2,2))
@test positions(ruleB, (10, 10)) ==
((8, 8), (9, 8), (10, 8), (11, 8), (12, 8),
(8, 9), (9, 9), (10, 9), (11, 9), (12, 9),
(8, 10), (9, 10), (11, 10), (12, 10),
(8, 11), (9, 11), (10, 11), (11, 11), (12, 11),
(8, 12), (9, 12), (10, 12), (11, 12), (12, 12))
end
@testset "radius" begin
init = (a=[1.0 2.0], b=[10.0 11.0])
ruleA = TestNeighborhoodRule{:a,:a}(Moore{3}(win7x7))
ruleB = TestSetNeighborhoodRule{Tuple{:b},Tuple{:b}}(Moore{2}(win5x5))
ruleset = Ruleset(ruleA, ruleB)
@test radius(ruleA) == 3
@test radius(ruleB) == 2
@test radius(ruleset) == (a=3, b=2)
@test radius(Ruleset()) == NamedTuple()
output = ArrayOutput(init; tspan=1:3)
sim!(output, ruleset)
# TODO make sure 2 radii can coexist
end
@testset "Convolution" begin
k = SA[1 0 1; 0 0 0; 1 0 1]
@test Convolution{:a}(k) == Convolution{:a,:a}(; neighborhood=Kernel(Window(1), k))
window = SA[1 0 0; 0 0 1; 0 0 1]
hood = Window{1,2,9,typeof(window)}(window)
rule = Convolution{:a,:a}(; neighborhood=Kernel(hood, k))
@test DynamicGrids.kernel(rule) === k
@test applyrule(nothing, rule, 0, (3, 3)) == k ⋅ window
output = ArrayOutput((a=init,); tspan=1:2)
sim!(output, rule)
end
@testset "SetNeighbors" begin
init = [0 1 0 0
0 0 0 0
0 0 0 0
0 1 0 0
0 0 1 0]
@test_throws ArgumentError SetNeighbors()
@testset "atomics" begin
rule = SetNeighbors(VonNeumann(1)) do data, hood, state, I
if state > 0
for pos in positions(hood, I)
add!(data, 1, pos...)
end
end
end
output = ArrayOutput(init; tspan=1:2)
for proc in hardware, opt in (NoOpt(), SparseOpt())
@testset "$(nameof(typeof(opt))) $(nameof(typeof(proc)))" begin
ref_output = [1 1 1 0
0 1 0 0
0 1 0 0
1 1 2 0
0 2 1 1]
sim!(output, rule, proc=proc, opt=opt)
@test output[2] == ref_output
end
end
end
@testset "setindex" begin
rule = SetNeighbors(VonNeumann(1)) do data, hood, state, I
state == 0 && return nothing
for pos in positions(hood, I)
data[pos...] = 1
end
end
output = ArrayOutput(init; tspan=1:2)
for proc in hardware, opt in (NoOpt(), SparseOpt())
@testset "$(nameof(typeof(opt))) $(nameof(typeof(proc)))" begin
ref_output = [1 1 1 0
0 1 0 0
0 1 0 0
1 1 1 0
0 1 1 1]
sim!(output, rule, proc=proc, opt=opt)
@test output[2] == ref_output
end
end
end
end
@testset "SetCell" begin
init = [0 1 0 0
0 0 0 0
0 0 0 0
0 1 0 0
0 0 1 0]
@testset "add!" begin
output = ArrayOutput(init; tspan=1:2)
rule = SetCell() do data, state, I
if state > 0
pos = I[1] - 2, I[2]
isinbounds(data, pos) && add!(first(data), 1, pos...)
end
end
for proc in hardware, opt in (NoOpt(), SparseOpt())
@testset "$(nameof(typeof(opt))) $(nameof(typeof(proc)))" begin
sim!(output, rule; proc=proc, opt=opt)
ref_out = [0 1 0 0
0 1 0 0
0 0 1 0
0 1 0 0
0 0 1 0]
@test output[2] == ref_out
end
end
end
@testset "setindex!" begin
output = ArrayOutput(init; tspan=1:2)
rule = SetCell() do data, state, I
if state > 0
pos = I[1] - 2, I[2]
isinbounds(data, pos) && (data[pos...] = 5)
end
end
sim!(output, rule)
@test output[2] == [0 1 0 0
0 5 0 0
0 0 5 0
0 1 0 0
0 0 1 0]
end
end
@testset "SetGrid" begin
@test_throws ArgumentError SetGrid()
rule = SetGrid() do r, w
w .*= 2
end
init = [0 1 0 0
0 0 0 0
0 0 0 0
0 1 0 0
0 0 1 0]
output = ArrayOutput(init; tspan=1:2)
for proc in hardware, opt in (NoOpt(), SparseOpt())
@testset "$(nameof(typeof(opt))) $(nameof(typeof(proc)))" begin
sim!(output, rule; proc=proc, opt=opt)
@test output[2] == [0 2 0 0
0 0 0 0
0 0 0 0
0 2 0 0
0 0 2 0]
end
end
end
struct AddOneRule{R,W} <: CellRule{R,W} end
DynamicGrids.applyrule(data, ::AddOneRule, state, args...) = state + 1
@testset "Rulset mask ignores false cells" begin
init = [0.0 4.0 0.0
0.0 5.0 8.0
3.0 6.0 0.0]
mask = Bool[0 1 0
0 1 1
1 1 0]
ruleset1 = Ruleset(AddOneRule{:_default_,:_default_}(); opt=NoOpt())
ruleset2 = Ruleset(AddOneRule{:_default_,:_default_}(); opt=SparseOpt())
output1 = ArrayOutput(init; tspan=1:3, mask=mask)
output2 = ArrayOutput(init; tspan=1:3, mask=mask)
sim!(output1, ruleset1)
sim!(output2, ruleset2)
@test output1[1] == output2[1] == [0.0 4.0 0.0
0.0 5.0 8.0
3.0 6.0 0.0]
@test output1[2] == output2[2] == [0.0 5.0 0.0
0.0 6.0 9.0
4.0 7.0 0.0]
@test output1[3] == output2[3] == [0.0 6.0 0.0
0.0 7.0 10.0
5.0 8.0 0.0]
end
# Single grid rules
struct TestRule{R,W} <: CellRule{R,W} end
applyrule(data, ::TestRule, state, index) = 0
@testset "A rule that returns zero gives zero outputs" begin
final = [0 0 0 0
0 0 0 0
0 0 0 0
0 0 0 0
0 0 0 0]
mask = nothing
rule = TestRule{:a,:a}()
for proc in (SingleCPU(), ThreadedCPU()), opt in (NoOpt(), SparseOpt())
@testset "$(nameof(typeof(opt))) $(nameof(typeof(proc)))" begin
ruleset = Ruleset(rule; opt=opt, proc=proc)
@test DynamicGrids.boundary(ruleset) === Remove()
@test DynamicGrids.opt(ruleset) === opt
@test DynamicGrids.proc(ruleset) === proc
@test DynamicGrids.cellsize(ruleset) === 1
@test DynamicGrids.timestep(ruleset) === nothing
@test DynamicGrids.ruleset(ruleset) === ruleset
ext = Extent(; init=(a=init,), tspan=1:1)
simdata = SimData(ext, ruleset)
# Test maprules components
rkeys, rgrids = _getreadgrids(rule, simdata)
wkeys, wgrids = _getwritegrids(rule, simdata)
@test rkeys == Val{:a}()
@test wkeys == Val{:a}()
newsimdata = @set simdata.grids = _combinegrids(rkeys, rgrids, wkeys, wgrids)
@test newsimdata.grids[1] isa WritableGridData
# Test type stability
T = Val{DynamicGrids.ruletype(rule)}()
@inferred maprule!(newsimdata, proc, opt, T, rule, rkeys, wkeys)
resultdata = maprule!(simdata, rule)
@test source(resultdata[:a]) == final
end
end
end
struct TestSetCell{R,W} <: SetCellRule{R,W} end
applyrule!(data, ::TestSetCell, state, index) = 0
@testset "A SetRule that returns zero does nothing" begin
rule = TestSetCell()
mask = nothing
for proc in (SingleCPU(), ThreadedCPU()), opt in (NoOpt(), SparseOpt())
@testset "$(nameof(typeof(opt))) $(nameof(typeof(proc)))" begin
ruleset = Ruleset(rule; opt=NoOpt())
# Test type stability
ext = Extent(; init=(_default_=init,), tspan=1:1)
simdata = SimData(ext, ruleset)
rkeys, rgrids = _getreadgrids(rule, simdata)
wkeys, wgrids = _getwritegrids(rule, simdata)
newsimdata = @set simdata.grids = _combinegrids(wkeys, wgrids, rkeys, rgrids)
T = Val{DynamicGrids.ruletype(rule)}()
@inferred maprule!(newsimdata, proc, opt, T, rule, rkeys, wkeys)
resultdata = maprule!(simdata, rule)
@test source(resultdata[:_default_]) == init
end
end
end
struct TestSetCellWrite{R,W} <: SetCellRule{R,W} end
applyrule!(data, ::TestSetCellWrite{R,W}, state, index) where {R,W} = add!(data[W], 1, index[1], 2)
@testset "A partial rule that writes to dest affects output" begin
init = [0 1 1 0
0 1 1 0
0 1 1 0
0 1 1 0
0 1 1 0]
final = [0 5 1 0;
0 5 1 0;
0 5 1 0;
0 5 1 0;
0 5 1 0]
for proc in hardware, opt in (NoOpt(), SparseOpt())
@testset "$(nameof(typeof(opt))) $(nameof(typeof(proc)))" begin
rule = TestSetCellWrite()
ruleset = Ruleset(rule; opt=opt, proc=proc)
ext = Extent(; init=(_default_=init,), tspan=1:1)
simdata = DynamicGrids._proc_setup(SimData(ext, ruleset));
resultdata = maprule!(simdata, rule);
@test Array(source(first(resultdata))) == final
end
end
end
struct TestCellTriple{R,W} <: CellRule{R,W} end
applyrule(data, ::TestCellTriple, state, index) = 3state
struct TestCellSquare{R,W} <: CellRule{R,W} end
applyrule(data, ::TestCellSquare, (state,), index) = state^2
@testset "Chained cell rules work" begin
init = [0 1 2 3;
4 5 6 7]
final = [0 9 36 81;
144 225 324 441]
for proc in hardware, opt in (NoOpt(), SparseOpt())
@testset "$(nameof(typeof(opt))) $(nameof(typeof(proc)))" begin
rule = Chain(TestCellTriple(), TestCellSquare())
ruleset = Ruleset(rule; opt=opt, proc=proc)
ext = Extent(; init=(_default_=init,), tspan=1:1)
simdata = DynamicGrids._proc_setup(SimData(ext, ruleset))
resultdata = maprule!(simdata, rule);
@test Array(source(first(resultdata))) == final
end
end
end
struct PrecalcRule{R,W,P} <: CellRule{R,W}
precalc::P
end
DynamicGrids.modifyrule(rule::PrecalcRule, simdata) = PrecalcRule(currenttime(simdata))
applyrule(data, rule::PrecalcRule, state, index) = rule.precalc[]
@testset "Rule precalculations work" begin
init = [1 1;
1 1]
out2 = [2 2;
2 2]
out3 = [3 3;
3 3]
for proc in hardware, opt in (NoOpt(), SparseOpt())
@testset "$(nameof(typeof(opt))) $(nameof(typeof(proc)))" begin
rule = PrecalcRule(1)
ruleset = Ruleset(rule; proc=proc, opt=opt)
output = ArrayOutput(init; tspan=1:3)
sim!(output, ruleset)
# Copy for GPU
@test output[2] == out2
@test output[3] == out3
end
end
end
# Multi grid rules
struct DoubleY{R,W} <: CellRule{R,W} end
applyrule(data, rule::DoubleY, (x, y), index) = y * 2
struct HalfX{R,W} <: CellRule{R,W} end
applyrule(data, rule::HalfX, x, index) = x, x * 0.5
struct Predation{R,W} <: CellRule{R,W} end
Predation(; prey=:prey, predator=:predator) =
Predation{Tuple{predator,prey},Tuple{prey,predator}}()
applyrule(data, ::Predation, (predators, prey), index) = begin
caught = 2predators
# Output order is the reverse of input to test that can work
prey - caught, predators + caught * 0.5
end
predation = Predation(; prey=:prey, predator=:predator)
@testset "Multi-grid keys are inferred" begin
@test _writekeys(predation) == (:prey, :predator)
@test _readkeys(predation) == (:predator, :prey)
@test keys(predation) == (:prey, :predator)
@inferred _writekeys(predation)
@inferred _readkeys(predation)
@inferred keys(predation)
end
@testset "Multi-grid rules work" begin
init = (prey=[10. 10.], predator=[1. 0.])
rules = DoubleY{Tuple{:predator,:prey},:prey}(), predation
output = ArrayOutput(init; tspan=1:3)
for proc in hardware, opt in (NoOpt(), SparseOpt())
@testset "$(nameof(typeof(opt))) $(nameof(typeof(proc)))" begin
sim!(output, rules; opt=opt, proc=proc)
@test output[2] == (prey=[18. 20.], predator=[2. 0.])
@test output[3] == (prey=[32. 40.], predator=[4. 0.])
end
end
end
@testset "Multi-grid rules work" begin
init = (prey=[10. 10.], predator=[0. 0.])
output = ArrayOutput(init; tspan=1:3)
for proc in hardware, opt in (NoOpt(), SparseOpt())
@testset "$(nameof(typeof(opt))) $(nameof(typeof(proc)))" begin
ruleset = Ruleset((HalfX{:prey,Tuple{:prey,:predator}}(),); opt=opt, proc=proc)
sim!(output, ruleset)
@test output[2] == (prey=[10. 10.], predator=[5. 5.])
@test output[3] == (prey=[10. 10.], predator=[5. 5.])
end
end
end
@testset "life with generic constructors" begin
@test Life(Moore(1), (1, 1), (5, 5)) ==
Life(; neighborhood=Moore(1), born=(1, 1), survive=(5, 5))
@test Life{:a,:b}(Moore(1), (7, 1), (5, 3)) ==
Life{:a,:b}(neighborhood=Moore(1), born=(7, 1), survive=(5, 3))
# Defaults
@test Life() == Life(
Moore(1),
Param(3, bounds=(0, 8)),
(Param(2, bounds=(0, 8)), Param(3, bounds=(0, 8)))
)
@test Life{:a,:b}() == Life{:a,:b}(
Moore(1),
Param(3, bounds=(0, 8)),
(Param(2, bounds=(0, 8)), Param(3, bounds=(0, 8)))
)
end
@testset "generic ConstructionBase compatability" begin
life = Life{:x,:y}(; neighborhood=Moore(2), born=(1, 1), survive=(2, 2))
@set! life.born = (5, 6)
@test life.born == (5, 6)
@test life.survive == (2, 2)
@test _readkeys(life) == :x
@test _writekeys(life) == :y
@test DynamicGrids.neighborhood(life) == Moore(2)
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 1495 | using DynamicGrids, Aqua, SafeTestsets
if VERSION >= v"1.5.0"
# Amibiguities are not owned by DynamicGrids
# Aqua.test_ambiguities([DynamicGrids, Base, Core])
Aqua.test_unbound_args(DynamicGrids)
Aqua.test_undefined_exports(DynamicGrids)
Aqua.test_project_extras(DynamicGrids)
# Aqua.test_stale_deps(DynamicGrids)
Aqua.test_deps_compat(DynamicGrids)
Aqua.test_project_toml_formatting(DynamicGrids)
end
@time @safetestset "generated" begin include("generated.jl") end
@time @safetestset "rules" begin include("rules.jl") end
@time @safetestset "neighborhoods" begin include("neighborhoods.jl") end
@time @safetestset "simulationdata" begin include("simulationdata.jl") end
@time @safetestset "utils" begin include("utils.jl") end
@time @safetestset "chain" begin include("chain.jl") end
@time @safetestset "condition" begin include("condition.jl") end
@time @safetestset "outputs" begin include("outputs.jl") end
@time @safetestset "transformed" begin include("transformed.jl") end
@time @safetestset "integration" begin include("integration.jl") end
@time @safetestset "objectgrids" begin include("objectgrids.jl") end
@time @safetestset "parametersources" begin include("parametersources.jl") end
@time @safetestset "show" begin include("show.jl") end
@time @safetestset "textconfig" begin include("textconfig.jl") end
# ImageMagick breaks in windows travis for some reason
if !Sys.iswindows()
@time @safetestset "image" begin include("image.jl") end
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 1211 | using DynamicGrids, Test, Dates
import DynamicGrids: Remove, NoOpt
life = Life()
@test occursin("Life", sprint((io, s) -> show(io, MIME"text/plain"(), s), life))
rs = Ruleset(;
rules=(Life(),),
timestep=Day(1),
boundary=Remove(),
opt=NoOpt(),
)
rs
@test occursin("Ruleset", sprint((io, s) -> show(io, MIME"text/plain"(), s), rs))
@test occursin("Life", sprint((io, s) -> show(io, MIME"text/plain"(), s), rs))
@test occursin(r"opt = .*NoOpt()", "nopt = DynamicGrids.NoOpt()")
@test occursin(r"boundary = .*Remove()", sprint((io, s) -> show(io, MIME"text/plain"(), s), rs))
rule1 = Cell{:a,:b}() do a
2a
end
@test occursin("Cell{:a,:b}", sprint((io, s) -> show(io, MIME"text/plain"(), s), rule1))
rule2 = Cell{Tuple{:b,:d},:c}() do b, d
b + d
end
@test occursin("Cell{Tuple{:b, :d},:c}", sprint((io, s) -> show(io, MIME"text/plain"(), s), rule2))
chain = Chain(rule1, rule2)
@test occursin("Chain{Tuple{:a, :b, :d},Tuple{:b, :c}}", sprint((io, s) -> show(io, MIME"text/plain"(), s), chain))
@test occursin("Cell{:a,:b}", sprint((io, s) -> show(io, MIME"text/plain"(), s), chain))
@test occursin("Cell{Tuple{:b, :d},:c}", sprint((io, s) -> show(io, MIME"text/plain"(), s), chain))
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 2757 | using DynamicGrids, OffsetArrays, Test, Dates
using DynamicGrids: initdata!, init, mask, boundary, source, dest,
sourcestatus, deststatus, gridsize, ruleset, grids, SimData, Extent,
_updatetime, WritableGridData, tspan, extent, optdata
inita = [0 1 1
0 1 1]
initb = [2 2 2
2 2 2]
initab = (a=inita, b=initb)
life = Life{:a,:a}()
tspan_ = DateTime(2001):Day(1):DateTime(2001, 2)
@testset "initdata!" begin
rs = Ruleset(life, timestep=Day(1); opt=SparseOpt());
ext = Extent(; init=initab, tspan=tspan_)
simdata = SimData(ext, rs)
@test simdata isa SimData
@test init(simdata) == initab
@test mask(simdata) === nothing
@test ruleset(simdata) === StaticRuleset(rs)
@test tspan(simdata) === tspan_
@test currentframe(simdata) === 1
@test first(simdata) === simdata[:a]
@test last(simdata) === simdata[:b]
@test boundary(simdata) === Remove()
@test gridsize(simdata) == (2, 3)
updated = _updatetime(simdata, 2)
@test currenttimestep(simdata) == Millisecond(86400000)
gs = grids(simdata)
grida = gs[:a]
gridb = gs[:b]
@test parent(source(grida)) == parent(dest(grida)) ==
[0 0 0 0 0
0 0 1 1 0
0 0 1 1 0
0 0 0 0 0
0 0 0 0 0
0 0 0 0 0]
wgrida = WritableGridData(grida)
@test parent(source(grida)) === parent(source(wgrida))
@test parent(dest(grida)) === parent(dest(wgrida))
@test parent(grida) == parent(wgrida) ==
[0 1 1
0 1 1]
@test sourcestatus(grida) == deststatus(grida) ==
[0 1 0 0
0 1 0 0
0 0 0 0
0 0 0 0]
@test parent(source(gridb)) == parent(dest(gridb)) ==
[2 2 2
2 2 2]
@test optdata(gridb) == optdata(gridb) == nothing
@test firstindex(grida) == 1
@test lastindex(grida) == 6
@test gridsize(grida) == (2, 3) == size(grida) == (2, 3)
@test axes(grida) == (1:2, 1:3)
@test ndims(grida) == 2
@test eltype(grida) == Int
output = ArrayOutput(initab; tspan=tspan_)
initdata!(simdata, output, extent(output), rs)
end
@testset "initdata! with :_default_" begin
initx = [1 0]
rs = Ruleset(Life())
output = ArrayOutput((_default_=initx,); tspan=tspan_)
simdata = SimData(output, rs)
simdata2 = initdata!(simdata, output, extent(output), rs)
@test keys(simdata2) == (:_default_,)
@test DynamicGrids.ruleset(simdata2) == DynamicGrids.StaticRuleset(rs)
@test DynamicGrids.init(simdata2)[:_default_] == [1 0]
@test DynamicGrids.source(simdata2[:_default_]) ==
OffsetArray([0 0 0 0
0 1 0 0
0 0 0 0
0 0 0 0
0 0 0 0], (0:4, 0:3))
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 662 | using DynamicGrids, FreeTypeAbstraction, Test
@testset "Fonts" begin
@test DynamicGrids.autofont() isa String
name = DynamicGrids.autofont()
face = FreeTypeAbstraction.findfont(name)
@testset "TextConfig accepts font as String" begin
@test name isa String
textconfig = TextConfig(; font=name)
@test textconfig.face isa FreeTypeAbstraction.FTFont
end
@testset "TextConfig accepts font as FTFont" begin
@test face isa FreeTypeAbstraction.FTFont
textconfig = TextConfig(; font=face)
@test textconfig.face === face
end
@test_throws ArgumentError TextConfig(; font="not_a_font")
@test_throws ArgumentError TextConfig(; font=:not_a_string)
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 4515 | using DynamicGrids, Test
# Reducing functions may return the original grid,
# not a copy, so we need to be careful with them.
@testset "Reducing functions over NamedTuple" begin
rule = Cell{:a}() do data, state, I
state + 10
end
@testset "single named grid" begin
init = (a=[1 3],)
transformed_output = TransformedOutput(sum, init; tspan=1:3)
@test length(transformed_output) == 3
@test transformed_output[1] == [1 3]
@test transformed_output[2] == [0 0]
@test transformed_output[3] == [0 0]
sim!(transformed_output, rule)
@test transformed_output[1] == [1 3]
@test transformed_output[2] == [11 13]
@test transformed_output[3] == [21 23]
end
@testset "multiple named grids" begin
init = (a=[1 3], b=[5 5],)
transformed_output = TransformedOutput(sum, init; tspan=1:3)
@test length(transformed_output) == 3
@test transformed_output[1] == [6 8]
@test transformed_output[2] == [0 0]
@test transformed_output[3] == [0 0]
sim!(transformed_output, rule)
@test transformed_output[1] == [6 8]
@test transformed_output[2] == [16 18]
@test transformed_output[3] == [26 28]
end
end
@testset "Reducing functions over Array" begin
rule = Cell() do data, state, I
state + 10.0
end
init = [1 3]
transformed_output = TransformedOutput(sum, init; tspan=1:3)
@test length(transformed_output) == 3
@test transformed_output[1] == 4
@test transformed_output[2] == 0
@test transformed_output[3] == 0
sim!(transformed_output, rule)
@test transformed_output[1] == 4
@test transformed_output[2] == 24
@test transformed_output[3] == 44
end
@testset "Padded grids" begin
init = Bool[
0 0 0 0 0 0 0
0 0 0 0 1 1 1
0 0 0 0 0 0 1
0 0 0 0 0 1 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
]
test2 = Bool[
0 0 0 0 0 1 0
0 0 0 0 0 1 1
0 0 0 0 1 0 1
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
]
test3 = Bool[
0 0 0 0 0 1 1
0 0 0 0 1 0 1
0 0 0 0 0 0 1
0 0 0 0 0 0 0
0 0 0 0 0 0 0
0 0 0 0 0 0 0
]
# Sum the last column in the grid
output = TransformedOutput(A -> sum(view(A, :, 7)), init; tspan=1:3)
sim!(output, Life())
output == [2, 2, 3]
# Sum the first row in the :a grid of the NamedTuple
output = TransformedOutput(gs -> sum(view(gs[:a], 1, :)), (a=init,); tspan=1:3)
sim!(output, Life{:a}())
output == [0, 1, 2]
end
@testset "Defining new function" begin
ruleAR = Cell() do data, state, I
state + 10.0
end
ruleNT = Cell{:a, :a}() do data, state, I
state + 10.0
end
# Defining 2 functions giving square of all single values
f_square(AR::AbstractArray) = map(x -> x^2, AR)
function f_square(NT::NamedTuple)
v = [map(x -> x^2, el) for el in values(NT)]
return NamedTuple{keys(NT)}(v)
end
@testset "Array" begin
init = [1 3]
f_square(init) == [1 9]
transformed_output = TransformedOutput(f_square, init; tspan=1:3)
sim!(transformed_output, ruleAR)
@test transformed_output[1] == [1 3] .^2
@test transformed_output[2] == ([1 3] .+ 10) .^2
@test transformed_output[3] == ([1 3] .+ 10 .+ 10) .^2
end
@testset "Single NamedTuple" begin
init = (a = [1 3],)
f_square(init) == (a = [ 1 9],)
transformed_output = TransformedOutput(f_square, init; tspan=1:3)
sim!(transformed_output, ruleNT)
@test transformed_output[1] == (a=[1 3] .^2 ,)
@test transformed_output[2] == (a=([1 3] .+ 10) .^2 ,)
@test transformed_output[3] == (a=([1 3] .+ 10 .+ 10) .^2 ,)
end
@testset "Multiple NamedTuple" begin
init = (a = [1 3], b = [2 5])
@test f_square(init) == (a = [1 9], b = [4 25])
transformed_output = TransformedOutput(f_square, init; tspan=1:3)
sim!(transformed_output, ruleNT)
@test transformed_output[1] == (a=[1 3] .^2 , b=[2 5] .^2)
@test transformed_output[2] == (a=([1 3] .+ 10) .^2 , b=[2 5] .^2)
@test transformed_output[3] == (a=([1 3] .+ 10 .+ 10) .^2 , b=[2 5] .^2)
end
end | DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | code | 3895 | using DynamicGrids, Test
using DynamicGrids: inbounds, isinbounds, _inbounds, _isinbounds, _cyclic_index,
SimData, _unwrap, ismasked
@testset "boundary boundary checks are working" begin
@testset "inbounds with Remove() returns index and false for an boundaryed index" begin
@test _inbounds(Remove(), (4, 5), 1, 1) == ((1,1), true)
@test _inbounds(Remove(), (4, 5), 2, 3) == ((2,3), true)
@test _inbounds(Remove(), (4, 5), 4, 5) == ((4,5), true)
@test _inbounds(Remove(), (4, 5), 0, 0) == ((0,0), false)
@test _inbounds(Remove(), (3, 2), 2, 3) == ((2,3), false)
@test _inbounds(Remove(), (1, 4), 2, 3) == ((2,3), false)
@test _inbounds(Remove(), (2, 3), 200, 300) == ((200,300), false)
@test _inbounds(Remove(), (4, 5), -3, -100) == ((-3,-100), false)
end
@testset "inbounds with Wrap() returns new index and true for an boundaryed index" begin
@test _inbounds(Wrap(), (10, 10), -2, 3) == ((8, 3), true)
@test _inbounds(Wrap(), (10, 10), 2, 0) == ((2, 10), true)
@test _inbounds(Wrap(), (10, 10), 22, 0) == ((2, 10), true)
@test _inbounds(Wrap(), (10, 10), -22, 0) == ((8, 10), true)
end
@testset "isinbounds" begin
@test _isinbounds((4, 5), 4, 5) == true
@test _isinbounds((2, 3), 200, 300) == false
@test _isinbounds((10, 10), -22, 0) == false
end
@testset "boundscheck objects" begin
output = ArrayOutput(zeros(Int, 10, 10); tspan=1:10)
sd = SimData(output.extent, Ruleset())
@test inbounds(sd, 5, 5) == ((5, 5), true)
@test inbounds(first(sd), 5, 5) == ((5, 5), true)
@test inbounds(sd, 12, 5) == ((12, 5), false)
sd_wrap = SimData(output.extent, Ruleset(; boundary=Wrap()))
@test inbounds(sd_wrap, 5, 5) == ((5, 5), true)
@test inbounds(sd_wrap, 12, 5) == ((2, 5), true)
@test inbounds(first(sd_wrap), 12, 5) == ((2, 5), true)
end
end
@testset "isinferred" begin
@testset "unstable conditional" begin
rule = let threshold = 20
Cell() do data, x, I
x > 1 ? 2 : 0.0
end
end
output = ArrayOutput(rand(Int, 10, 10); tspan=1:10)
@test_throws ErrorException isinferred(output, rule)
end
@testset "return type" begin
rule = Neighbors{:a,:a}(Moore{1}(zeros(Bool, 3, 3))) do data, hood, x, I
round(Int, x + sum(hood))
end
output = ArrayOutput((a=rand(Int, 10, 10),); tspan=1:10)
@test isinferred(output, rule)
output = ArrayOutput((a=rand(Bool, 10, 10),); tspan=1:10)
@test_throws ErrorException isinferred(output, rule)
end
@testset "let blocks" begin
a = 0.7
rule = SetCell() do data, x, I
add!(first(data), round(Int, a + x), I...)
end
output = ArrayOutput(zeros(Int, 10, 10); tspan=1:10)
@test_throws ErrorException isinferred(output, Ruleset(rule))
a = 0.7
rule = let a = a
SetCell() do data, x, I
add!(first(data), round(Int, a), I...)
end
end
output = ArrayOutput(zeros(Int, 10, 10); tspan=1:10)
@test isinferred(output, Ruleset(rule))
end
end
@testset "ismasked" begin
output = ArrayOutput(zeros(2, 2); mask=Bool[1 0; 0 1], tspan=1:10)
sd = SimData(output.extent, Ruleset())
@test ismasked(sd, 1, 2) == true
@test ismasked(sd, 2, 2) == false
output_nomask = ArrayOutput(zeros(2, 2); tspan=1:10)
sd = SimData(output_nomask.extent, Ruleset())
@test ismasked(sd, 1, 2) == false
end
@testset "unwrap" begin
@test _unwrap(1) == 1
@test _unwrap(Val(:a)) == :a
@test _unwrap(Aux(:a)) == :a
@test _unwrap(Grid(:a)) == :a
@test _unwrap(Aux{:x}) == :x
@test _unwrap(Grid{:x}) == :x
@test _unwrap(Val{:x}) == :x
end
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | docs | 14929 | 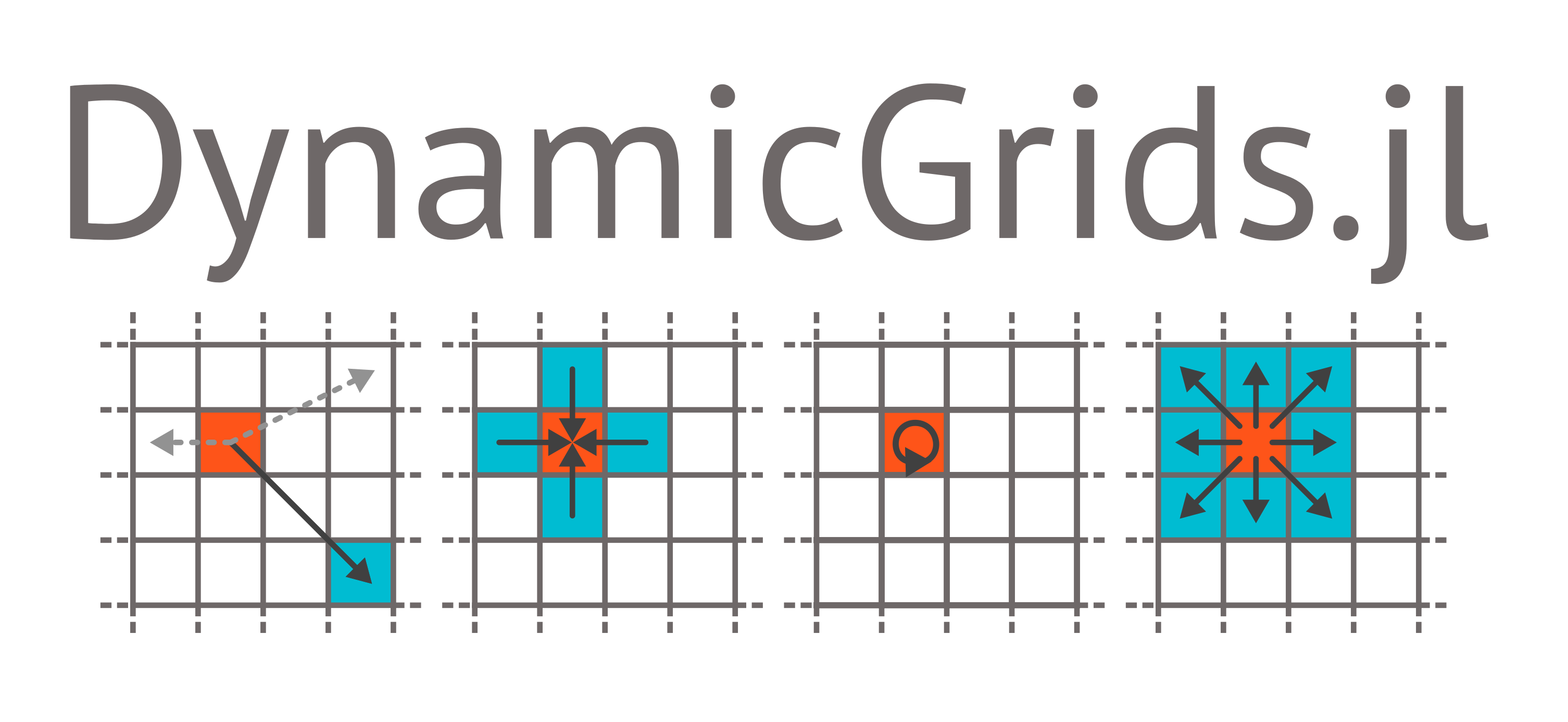
[](https://cesaraustralia.github.io/DynamicGrids.jl/stable)
[](https://cesaraustralia.github.io/DynamicGrids.jl/dev)
[](https://github.com/cesaraustralia/DynamicGrids.jl/actions/workflows/ci.yml)
[](http://codecov.io/github/cesaraustralia/DynamicGrids.jl?branch=master)
[](https://github.com/JuliaTesting/Aqua.jl)
DynamicGrids is a generalised framework for building high-performance grid-based
spatial simulations, including cellular automata, but also allowing a wider
range of behaviours like random jumps and interactions between multiple grids.
It is extended by [Dispersal.jl](https://github.com/cesaraustralia/Dispersal.jl)
for modelling organism dispersal processes.
[DynamicGridsGtk.jl](https://github.com/cesaraustralia/DynamicGridsGtk.jl) provides a simple live
interface, while [DynamicGridsInteract.jl](https://github.com/cesaraustralia/DynamicGridsInteract.jl)
also has live control over model parameters while the simulation runs: real-time visual feedback for
manual parametrisation and model exploration.
DynamicGrids can run rules on single CPUs, threaded CPUs, and on CUDA GPUs.
Simulation run-time is usually measured in fractions of a second.
*A dispersal simulation with quarantine interactions, using Dispersal.jl, custom rules and the
GtkOuput from [DynamicGridsGtk](https://github.com/cesaraustralia/DynamicGridsGtk.jl).
Note that this is indicative of the real-time frame-rate on a laptop.*
A DynamicGrids.jl simulation is run with a script like this one
running the included game of life model `Life()`:
```julia
using DynamicGrids, Crayons
init = rand(Bool, 150, 200)
output = REPLOutput(init; tspan=1:200, fps=30, color=Crayon(foreground=:red, background=:black, bold=true))
sim!(output, Life())
# Or define it from scratch (yes this is actually the whole implementation!)
life = Neighbors(Moore(1)) do data, hood, state, I
born_survive = (false, false, false, true, false, false, false, false, false),
(false, false, true, true, false, false, false, false, false)
born_survive[state + 1][sum(hood) + 1]
end
sim!(output, life)
```

*A game of life simulation being displayed directly in a terminal.*
# Concepts
The framework is highly customisable, but there are some central ideas that define
how a simulation works: *grids*, *rules*, and *outputs*.
## Grids
Simulations run over one or many grids, derived from `init` of a single
`AbstractArray` or a `NamedTuple` of multiple `AbstractArray`. Grids (`GridData`
types) are, however not a single array but both source and destination arrays,
to maintain independence between cell reads and writes where required. These may
be padded or otherwise altered for specific performance optimisations. However,
broadcasted `getindex` operations are guaranteed to work on them as if the grid
is a regular array. This may be useful running simulations manually with
`step!`.
### Grid contents
Often grids contain simple values of some kind of `Number`, but other types are
possible, such as `SArray`, `FieldVector` or other custom structs. Grids are
updated by `Rule`s that are run for every cell, at every timestep.
NOTE: Grids of mutable objects (e.g `Array` or any `mutable struct` have
undefined behaviour. DynamicGrids.jl does not `deepcopy` grids between frames as
it is expensive, so successive frames will contain the same objects. Mutable
objects will not work at all on GPUs, and are relatively slow on CPUs. Instead,
use regular immutable structs and `StaticArrays.jl` if you need arrays. Update
them using `@set` from Setfield.jl or Accessors.jl, and generally use functional
programming approaches over object-oriented ones.
### Init
The `init` grid/s contain whatever initialisation data is required to start
a simulation: the array type, size and element type, as well as providing the
initial conditions:
```julia
init = rand(Float32, 100, 100)
```
An `init` grid can be attached to an `Output`:
```julia
output = ArrayOutput(init; tspan=1:100)
```
or passed in to `sim!`, where it will take preference over the `init`
attached to the `Output`, but must be the same type and size:
```julia
sim!(output, ruleset; init=init)
```
For multiple grids, `init` is a `NamedTuple` of equal-sized arrays
matching the names used in each `Ruleset` :
```julia
init = (predator=rand(100, 100), prey=(rand(100, 100))
```
Handling and passing of the correct grids to a `Rule` is automated by
DynamicGrids.jl, as a no-cost abstraction. `Rule`s specify which grids they
require in what order using the first two (`R` and `W`) type parameters.
Dimensional or spatial `init` grids from
[DimensionalData.jl](https://github.com/rafaqz/DimensionalData.jl) or
[GeoData.jl](https://github.com/rafaqz/GeoData.jl) will propagate through the
model to return output with explicit dimensions. This will plot correctly as a
map using [Plots.jl](https://github.com/JuliaPlots/Plots.jl), to which shape
files and observation points can be easily added.
### Non-Number Grids
Grids containing custom and non-`Number` types are possible, with some caveats.
They must define `Base.zero` for their element type, and should be a bitstype for performance.
Tuple does not define `zero`. `Array` is not a bitstype, and does not define `zero`.
`SArray` from StaticArrays.jl is both, and can be used as the contents of a grid.
Custom structs that defne `zero` should also work.
However, for any multi-values grid element type, you will need to define a method of
`DynamicGrids.to_rgb` that returns an `ARGB32` for them to work in `ImageOutput`s, and
`isless` for the `REPLoutput` to work. A definition for multiplication by a scalar `Real`
and addition are required to use `Convolution` kernels.
## Rules
Rules hold the parameters for running a simulation, and are applied in
`applyrule` method that is called for each of the active cells in the grid.
Rules come in a number of flavours (outlined in the
[docs](https://cesaraustralia.github.io/DynamicGrids.jl/stable/#Rules-1)). This
allows using specialised methods for different types of rules, ecoding assumtions
about their behaviours that can greatly improve performance through more efficient
use of caches and parallelisation. Rules can be collected in a `Ruleset`, with some
additional arguments to control the simulation:
```julia
ruleset = Ruleset(Life(2, 3); opt=SparseOpt(), proc=CuGPU())
```
Multiple rules can be combined in a `Ruleset` or simply passed to `sim!` directly. Each rule
will be run for the whole grid, in sequence, using appropriate optimisations depending
on the parent types of each rule:
```julia
ruleset = Ruleset(rule1, rule2; timestep=Day(1), opt=SparseOpt(), proc=ThreadedCPU())
```
## Output
[Outputs](https://cesaraustralia.github.io/DynamicGrids.jl/stable/#Output-1)
are ways of storing or viewing a simulation. They can be used
interchangeably depending on your needs: `ArrayOutput` is a simple storage
structure for high performance-simulations. As with most outputs, it is
initialised with the `init` array, but in this case it also requires the number
of simulation frames to preallocate before the simulation runs.
```julia
output = ArrayOutput(init; tspan=1:10)
```
The `REPLOutput` shown above is a `GraphicOutput` that can be useful for checking a
simulation when working in a terminal or over ssh:
```julia
output = REPLOutput(init; tspan=1:100)
```
`ImageOutput` is the most complex class of outputs, allowing full color visual
simulations using ColorSchemes.jl. It can also display multiple grids using color
composites or layouts, as shown above in the quarantine simulation.
[DynamicGridsInteract.jl](https://github.com/cesaraustralia/DynamicGridsInteract.jl)
provides simulation interfaces for use in Juno, Jupyter, web pages or electron
apps, with live interactive control over parameters, using
[ModelParameters.jl](https://github.com/rafaqz/ModelParameters.jl).
[DynamicGridsGtk.jl](https://github.com/cesaraustralia/DynamicGridsGtk.jl) is a
simple graphical output for Gtk. These packages are kept separate to avoid
dependencies when being used in non-graphical simulations.
Outputs are also easy to write, and high performance applications may benefit
from writing a custom output to reduce memory use, or using `TransformedOuput`.
Performance of DynamicGrids.jl is dominated by cache interactions, so reducing
memory use has positive effects.
## Example: Forest Fire
This example implements the classic stochastic forest fire model in a few
different ways, and benchmarks them. Note you will need ImageMagick.jl
installed for `.gif` output to work.
First we will define a Forest Fire algorithm that sets the current cell to
burning, if a neighbor is burning. Dead cells can come back to life, and living
cells can spontaneously catch fire:
```julia
using DynamicGrids, ColorSchemes, Colors, BenchmarkTools
const DEAD, ALIVE, BURNING = 1, 2, 3
neighbors_rule = let prob_combustion=0.0001, prob_regrowth=0.01
Neighbors(Moore(1)) do data, neighborhood, cell, I
if cell == ALIVE
if BURNING in neighborhood
BURNING
else
rand() <= prob_combustion ? BURNING : ALIVE
end
elseif cell == BURNING
DEAD
else
rand() <= prob_regrowth ? ALIVE : DEAD
end
end
end
# Set up the init array and output (using a Gtk window)
init = fill(ALIVE, 400, 400)
output = GifOutput(init;
filename="forestfire.gif",
tspan=1:200,
fps=25,
minval=DEAD, maxval=BURNING,
scheme=ColorSchemes.rainbow,
zerocolor=RGB24(0.0)
)
# Run the simulation, which will save a gif when it completes
sim!(output, neighbors_rule)
```

Timing the simulation for 200 steps, the performance is quite good. This
particular CPU has six cores, and we get a 5.25x speedup by using all of them,
which indicates good scaling:
```julia
bench_output = ResultOutput(init; tspan=1:200)
julia>
@btime sim!($bench_output, $neighbors_rule);
477.183 ms (903 allocations: 2.57 MiB)
julia> @btime sim!($bench_output, $neighbors_rule; proc=ThreadedCPU());
91.321 ms (15188 allocations: 4.07 MiB)
```
We can also _invert_ the algorithm, setting cells in the neighborhood to burning
if the current cell is burning, by using the `SetNeighbors` rule:
```julia
setneighbors_rule = let prob_combustion=0.0001, prob_regrowth=0.01
SetNeighbors(Moore(1)) do data, neighborhood, cell, I
if cell == DEAD
if rand() <= prob_regrowth
data[I...] = ALIVE
end
elseif cell == BURNING
for pos in positions(neighborhood, I)
if data[pos...] == ALIVE
data[pos...] = BURNING
end
end
data[I...] = DEAD
elseif cell == ALIVE
if rand() <= prob_combustion
data[I...] = BURNING
end
end
end
end
```
_Note: we are not using `add!`, instead we just set the grid value directly.
This usually risks errors if multiple cells set different values. Here they
only ever set a currently living cell to burning in the next timestep. It doesn't
matter if this happens multiple times, the result is the same._
And in this case (a fairly sparse simulation), this rule is faster:
```julia
julia> @btime sim!($bench_output, $setneighbors_rule);
261.969 ms (903 allocations: 2.57 MiB)
julia> @btime sim!($bench_output, $setneighbors_rule; proc=ThreadedCPU());
65.489 ms (7154 allocations: 3.17 MiB)
```
But the scaling is not quite as good, at 3.9x for 6 cores. The first
method may be better on a machine with a lot of cores.
Last, we slightly rewrite these rules for GPU, as `rand` was not available
within a GPU kernel. It is now, but it turns out that this method is faster!
and interesting to demonstrate using multiple grids and `SetGrid`.
This way we call `CUDA.rand!` on the entire parent array of the `:rand` grid,
using a `SetGrid` rule:
```julia
using CUDAKernels, CUDA
randomiser = SetGrid{Tuple{},:rand}() do randgrid
CUDA.rand!(parent(randgrid))
end
```
Now we define a Neighbors version for GPU, using the `:rand` grid values
instead of `rand()`:
```julia
neighbors_gpu = let prob_combustion=0.0001, prob_regrowth=0.01
Neighbors{Tuple{:ff,:rand},:ff}(Moore(1)) do data, neighborhood, (cell, rand), I
if cell == ALIVE
if BURNING in neighborhood
BURNING
else
rand <= prob_combustion ? BURNING : ALIVE
end
elseif cell == BURNING
DEAD
else
rand <= prob_regrowth ? ALIVE : DEAD
end
end
end
```
And a SetNeighbors version for GPU:
```julia
setneighbors_gpu = let prob_combustion=0.0001, prob_regrowth=0.01
SetNeighbors{Tuple{:ff,:rand},:ff}(Moore(1)) do data, neighborhood, (cell, rand), I
if cell == DEAD
if rand <= prob_regrowth
data[:ff][I...] = ALIVE
end
elseif cell == BURNING
for pos in positions(neighborhood, I)
if data[:ff][pos...] == ALIVE
data[:ff][pos...] = BURNING
end
end
data[:ff][I...] = DEAD
elseif cell == ALIVE
if rand <= prob_combustion
data[:ff][I...] = BURNING
end
end
end
end
```
Now benchmark both version on a GTX 1080 GPU. Despite the overhead of reading and
writing two grids, this turns out to be even faster again:
```julia
bench_output_rand = ResultOutput((ff=init, rand=zeros(size(init))); tspan=1:200)
julia> @btime sim!($bench_output_rand, $randomiser, $neighbors_gpu; proc=CuGPU());
30.621 ms (186284 allocations: 17.19 MiB)
julia> @btime sim!($bench_output_rand, $randomiser, $setneighbors_gpu; proc=CuGPU());
22.685 ms (147339 allocations: 15.61 MiB)
```
That is, we are running the rule at a rate of _1.4 billion times per second_.
These timings could be improved (maybe 10-20%) by using grids of `Int32` or
`Int16` to use less memory and cache. But we will stop here.
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.21.3 | a99984c15928d6579f670c53077e0f937b378e8a | docs | 6818 | # DynamicGrids
```@docs
DynamicGrids
```
## Running simulations
```@docs
sim!
resume!
step!
```
## Rulesets
```@docs
AbstractRuleset
Ruleset
```
## Options/Flags
### Boundary conditions
```@docs
BoundaryCondition
Wrap
Remove
```
### Hardware selection
```@docs
DynamicGrids.Processor
DynamicGrids.CPU
SingleCPU
ThreadedCPU
DynamicGrids.GPU
CuGPU
CPUGPU
```
### Performance optimisation
```@docs
PerformanceOpt
NoOpt
SparseOpt
```
## Rules
```@docs
Rule
DynamicGrids.SetRule
```
### CellRule
```@docs
CellRule
Cell
CopyTo
```
### NeighborhoodRule
```@docs
NeighborhoodRule
Neighbors
Convolution
Life
```
### SetCellRule
```@docs
SetCellRule
SetCell
```
### SetNeighborhoodRule
```@docs
SetNeighborhoodRule
SetNeighbors
```
### SetGridRule
```@docs
SetGridRule
SetGrid
```
### Rule wrappers
```@docs
RuleWrapper
Chain
RunIf
RunAt
```
### Parameter sources
```@docs
ParameterSource
Aux
Grid
DynamicGrids.AbstractDelay
Delay
Frame
Lag
```
### Custom Rule interface and helpers
```@docs
DynamicGrids.applyrule
DynamicGrids.applyrule!
DynamicGrids.modifyrule
isinferred
```
### Methods and objects for use in `applyrule` and/or `modifyrule`
```@docs
get
DynamicGrids.isinbounds
DynamicGrids.inbounds
DynamicGrids.ismasked
DynamicGrids.init
DynamicGrids.aux
DynamicGrids.mask
DynamicGrids.tspan
DynamicGrids.timestep
DynamicGrids.currenttimestep
DynamicGrids.currenttime
DynamicGrids.currentframe
DynamicGrids.AbstractSimData
DynamicGrids.SimData
DynamicGrids.RuleData
DynamicGrids.GridData
DynamicGrids.ReadableGridData
DynamicGrids.WritableGridData
DynamicGrids.AbstractSimSettings
DynamicGrids.SimSettings
```
## Neighborhoods
```@docs
Neighborhood
Moore
VonNeumann
Window
DynamicGrids.AbstractPositionalNeighborhood
Positional
LayeredPositional
```
### Methods for use with neighborhood rules and neighborhoods
```@docs
neighborhood
radius
distances
```
Useful with [`NeighborhoodRule`](@ref):
```@docs
neighbors
```
Useful with [`SetNeighborhoodRule`](@ref):
```@docs
positions
offsets
```
### Convolution kernel neighborhoods
```@docs
AbstractKernelNeighborhood
Kernel
kernel
kernelproduct
```
### Low level use of neighborhoods
```@docs
DynamicGrids.Neighborhoods.readwindow
DynamicGrids.Neighborhoods.unsafe_readwindow
DynamicGrids.Neighborhoods.updatewindow
DynamicGrids.Neighborhoods.unsafe_updatewindow
DynamicGrids.Neighborhoods.pad_axes
DynamicGrids.Neighborhoods.unpad_axes
```
### Generic neighborhood applicators
These can be used without the full simulation mechanisms, like `broadcast`.
```@docs
DynamicGrids.Neighborhoods.broadcast_neighborhood
DynamicGrids.Neighborhoods.broadcast_neighborhood!
```
## Atomic methods for SetCellRule and SetNeighborhoodRule
Using these methods to modify grid values ensures cell independence,
and also prevent race conditions with [`ThreadedCPU`](@ref) or [`CuGPU`].
```@docs
add!
sub!
min!
max!
and!
or!
xor!
```
## Output
### Output Types and Constructors
```@docs
Output
ArrayOutput
ResultOutput
TransformedOutput
GraphicOutput
REPLOutput
ImageOutput
GifOutput
```
### Renderers
```@docs
Renderer
DynamicGrids.SingleGridRenderer
Image
DynamicGrids.MultiGridRenderer
Layout
SparseOptInspector
```
### Color schemes
Schemes from Colorschemes.jl can be used for the `scheme` argument to `ImageOutput`,
`Renderer`s. `Greyscale` control over the band of grey used, and is very fast.
`ObjectScheme` is the default.
```@docs
ObjectScheme
Greyscale
```
### Text labels
```@docs
TextConfig
```
### Saving gifs
```@docs
savegif
```
### `Output` interface
These are used for defining your own outputs and `GridProcessors`,
not for general scripting.
```@docs
DynamicGrids.AbstractExtent
DynamicGrids.Extent
DynamicGrids.extent
DynamicGrids.isasync
DynamicGrids.storeframe!
DynamicGrids.isrunning
DynamicGrids.isshowable
DynamicGrids.isstored
DynamicGrids.initialise!
DynamicGrids.finalise!
DynamicGrids.frameindex
```
### `GraphicOutput` interface
Also includes `Output` interface.
```@docs
DynamicGrids.GraphicConfig
DynamicGrids.graphicconfig
DynamicGrids.fps
DynamicGrids.setfps!
DynamicGrids.showframe
DynamicGrids.initialisegraphics
DynamicGrids.finalisegraphics
```
### `ImageOutput` components and interface
Also uses `Output` and `GraphicOutput` interfaces.
```@docs
DynamicGrids.ImageConfig
DynamicGrids.imageconfig
DynamicGrids.showimage
DynamicGrids.render!
DynamicGrids.to_rgb
```
## Custom grid element types
It is common to use `Bool`, `Int` or `Float64` as the contents of a grid.
But a range of object types can be used if they meet the interface criteria.
Immutable, `isbits` objects are usually better and the only type officially to
work - as they are loaded directly in the simulation. Mutable objects,
especially containing pointers, may lead to incorrect stored results, and wont
work at all on GPUs.
Methods to define are:
- `zero`: define zero of the object type
- `oneunit`: define one of the object type
- `isless`: define comparison between two of the objects
- `*`: multiplication by a `Real` scalar.
- `/`: division by a `Real` scalar.
- `+`: addition to another object of the same type
- `-`: subtraction from another object of the same type
- `to_rgb`: return and `ARGB32` to visualise the object as a pixel
In this example we define a struct with two fields. You will need to determine the
correct behaviours for your own types, but hopefully this will get you started.
```julia
struct MYStruct{A,B}
a::A
b::B
end
Base.isless(a::MyStruct, b::MyStruct) = isless(a.a, b.a)
Base.zero(::Type{<:MyStruct{T1,T2}}) where {T1,T2} = MyStruct(zero(T1), zero(T2))
Base.oneunit(::Type{<:MyStruct{T1,T2}}) where {T1,T2} = MyStruct(one(T1), one(T2))
Base.:*(x::MyStruct, x::Number) = MyStruct(x.a * x, x.b * x)
Base.:*(x::Number, x::MyStruct) = MyStruct(x * x.a, x * x.b)
Base.:/(x::MyStruct, x::Number) = MyStruct(x.a / x, x.b / x)
Base.:+(x1::MyStruct, x2::MyStruct) = MyStruct(x1.a + x2.a, x1.b + x2.b)
Base.:-(x1::MyStruct, x2::MyStruct) = MyStruct(x1.a - x2.a, x1.b - x2.b)
```
To generate rgb colors for an `ImageOuput`, you must define `to_rgb`,
at least for the default `ObjectScheme`, but this can also be done for other
schemes such as ColorSchemes.jl, or `GreyScale`, by calling `get` on the scheme and a
`Real` value. Note that the objects will be normalised to values between zero and one
by `minval` and `maxval` scalars prior to this, using the division operators defined
above. It is preferable to use `minval` and `maxval` over normalising in `to_rgb` -
as this will not be as flexible for scripting.
```julia
DynamicGrids.to_rgb(::ObjectScheme, obj::MyStruct) = ARGB32(obj.a, obj.b, 0)
DynamicGrids.to_rgb(scheme, obj::MyStruct) = get(scheme, obj.a)
```
See the `test/objectgrids.jl` tests for more details on using complex objects in grids.
| DynamicGrids | https://github.com/cesaraustralia/DynamicGrids.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | code | 280 | push!(LOAD_PATH,"../src/")
using ExifViewer
using Documenter
DocMeta.setdocmeta!(ExifViewer, :DocTestSetup, :(using ExifViewer); recursive=true)
makedocs(;
modules=[ExifViewer],
sitename="ExifViewer.jl",
)
deploydocs(;
repo="github.com/JuliaImages/ExifViewer.jl",
) | ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | code | 484 | using Clang.JLLEnvs
using Clang.Generators
using libexif_jll
include_dir = normpath(libexif_jll.artifact_dir, "include")
prefix = "/libexif/"
headers = [
joinpath(include_dir * prefix, header) for
header in readdir(include_dir * prefix) if endswith(header, ".h")
]
options = load_options(joinpath(@__DIR__, "generator.toml"))
args = get_default_args()
push!(args, "-I$include_dir")
push!(args, "--include=stdint.h")
ctx = create_context(headers, args, options)
build!(ctx) | ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | code | 25061 | module LibExif
using libexif_jll
export libexif_jll
@enum ExifByteOrder::UInt32 begin
EXIF_BYTE_ORDER_MOTOROLA = 0
EXIF_BYTE_ORDER_INTEL = 1
end
function exif_byte_order_get_name(order)
ccall((:exif_byte_order_get_name, libexif), Ptr{Cchar}, (ExifByteOrder,), order)
end
mutable struct _ExifContentPrivate end
ExifContentPrivate = _ExifContentPrivate
mutable struct _ExifContent
entries::Ptr{Ptr{Cvoid}} # entries::Ptr{Ptr{ExifEntry}}
count::Cuint
parent::Ptr{Cvoid} # parent::Ptr{ExifData}
priv::Ptr{ExifContentPrivate}
end
function Base.getproperty(x::_ExifContent, f::Symbol)
f === :entries && return Ptr{Ptr{ExifEntry}}(getfield(x, f))
f === :parent && return Ptr{ExifData}(getfield(x, f))
return getfield(x, f)
end
ExifContent = _ExifContent
@enum ExifIfd::UInt32 begin
EXIF_IFD_0 = 0
EXIF_IFD_1 = 1
EXIF_IFD_EXIF = 2
EXIF_IFD_GPS = 3
EXIF_IFD_INTEROPERABILITY = 4
EXIF_IFD_COUNT = 5
end
function exif_content_get_ifd(c)
ccall((:exif_content_get_ifd, libexif), ExifIfd, (Ptr{ExifContent},), c)
end
@enum ExifTag::UInt32 begin
EXIF_TAG_INTEROPERABILITY_INDEX = 1
EXIF_TAG_INTEROPERABILITY_VERSION = 2
EXIF_TAG_NEW_SUBFILE_TYPE = 254
EXIF_TAG_IMAGE_WIDTH = 256
EXIF_TAG_IMAGE_LENGTH = 257
EXIF_TAG_BITS_PER_SAMPLE = 258
EXIF_TAG_COMPRESSION = 259
EXIF_TAG_PHOTOMETRIC_INTERPRETATION = 262
EXIF_TAG_FILL_ORDER = 266
EXIF_TAG_DOCUMENT_NAME = 269
EXIF_TAG_IMAGE_DESCRIPTION = 270
EXIF_TAG_MAKE = 271
EXIF_TAG_MODEL = 272
EXIF_TAG_STRIP_OFFSETS = 273
EXIF_TAG_ORIENTATION = 274
EXIF_TAG_SAMPLES_PER_PIXEL = 277
EXIF_TAG_ROWS_PER_STRIP = 278
EXIF_TAG_STRIP_BYTE_COUNTS = 279
EXIF_TAG_X_RESOLUTION = 282
EXIF_TAG_Y_RESOLUTION = 283
EXIF_TAG_PLANAR_CONFIGURATION = 284
EXIF_TAG_RESOLUTION_UNIT = 296
EXIF_TAG_TRANSFER_FUNCTION = 301
EXIF_TAG_SOFTWARE = 305
EXIF_TAG_DATE_TIME = 306
EXIF_TAG_ARTIST = 315
EXIF_TAG_WHITE_POINT = 318
EXIF_TAG_PRIMARY_CHROMATICITIES = 319
EXIF_TAG_SUB_IFDS = 330
EXIF_TAG_TRANSFER_RANGE = 342
EXIF_TAG_JPEG_PROC = 512
EXIF_TAG_JPEG_INTERCHANGE_FORMAT = 513
EXIF_TAG_JPEG_INTERCHANGE_FORMAT_LENGTH = 514
EXIF_TAG_YCBCR_COEFFICIENTS = 529
EXIF_TAG_YCBCR_SUB_SAMPLING = 530
EXIF_TAG_YCBCR_POSITIONING = 531
EXIF_TAG_REFERENCE_BLACK_WHITE = 532
EXIF_TAG_XML_PACKET = 700
EXIF_TAG_RELATED_IMAGE_FILE_FORMAT = 4096
EXIF_TAG_RELATED_IMAGE_WIDTH = 4097
EXIF_TAG_RELATED_IMAGE_LENGTH = 4098
EXIF_TAG_IMAGE_DEPTH = 32997
EXIF_TAG_CFA_REPEAT_PATTERN_DIM = 33421
EXIF_TAG_CFA_PATTERN = 33422
EXIF_TAG_BATTERY_LEVEL = 33423
EXIF_TAG_COPYRIGHT = 33432
EXIF_TAG_EXPOSURE_TIME = 33434
EXIF_TAG_FNUMBER = 33437
EXIF_TAG_IPTC_NAA = 33723
EXIF_TAG_IMAGE_RESOURCES = 34377
EXIF_TAG_EXIF_IFD_POINTER = 34665
EXIF_TAG_INTER_COLOR_PROFILE = 34675
EXIF_TAG_EXPOSURE_PROGRAM = 34850
EXIF_TAG_SPECTRAL_SENSITIVITY = 34852
EXIF_TAG_GPS_INFO_IFD_POINTER = 34853
EXIF_TAG_ISO_SPEED_RATINGS = 34855
EXIF_TAG_OECF = 34856
EXIF_TAG_TIME_ZONE_OFFSET = 34858
EXIF_TAG_SENSITIVITY_TYPE = 34864
EXIF_TAG_STANDARD_OUTPUT_SENSITIVITY = 34865
EXIF_TAG_RECOMMENDED_EXPOSURE_INDEX = 34866
EXIF_TAG_ISO_SPEED = 34867
EXIF_TAG_ISO_SPEEDLatitudeYYY = 34868
EXIF_TAG_ISO_SPEEDLatitudeZZZ = 34869
EXIF_TAG_EXIF_VERSION = 36864
EXIF_TAG_DATE_TIME_ORIGINAL = 36867
EXIF_TAG_DATE_TIME_DIGITIZED = 36868
EXIF_TAG_OFFSET_TIME = 36880
EXIF_TAG_OFFSET_TIME_ORIGINAL = 36881
EXIF_TAG_OFFSET_TIME_DIGITIZED = 36882
EXIF_TAG_COMPONENTS_CONFIGURATION = 37121
EXIF_TAG_COMPRESSED_BITS_PER_PIXEL = 37122
EXIF_TAG_SHUTTER_SPEED_VALUE = 37377
EXIF_TAG_APERTURE_VALUE = 37378
EXIF_TAG_BRIGHTNESS_VALUE = 37379
EXIF_TAG_EXPOSURE_BIAS_VALUE = 37380
EXIF_TAG_MAX_APERTURE_VALUE = 37381
EXIF_TAG_SUBJECT_DISTANCE = 37382
EXIF_TAG_METERING_MODE = 37383
EXIF_TAG_LIGHT_SOURCE = 37384
EXIF_TAG_FLASH = 37385
EXIF_TAG_FOCAL_LENGTH = 37386
EXIF_TAG_SUBJECT_AREA = 37396
EXIF_TAG_TIFF_EP_STANDARD_ID = 37398
EXIF_TAG_MAKER_NOTE = 37500
EXIF_TAG_USER_COMMENT = 37510
EXIF_TAG_SUB_SEC_TIME = 37520
EXIF_TAG_SUB_SEC_TIME_ORIGINAL = 37521
EXIF_TAG_SUB_SEC_TIME_DIGITIZED = 37522
EXIF_TAG_XP_TITLE = 40091
EXIF_TAG_XP_COMMENT = 40092
EXIF_TAG_XP_AUTHOR = 40093
EXIF_TAG_XP_KEYWORDS = 40094
EXIF_TAG_XP_SUBJECT = 40095
EXIF_TAG_FLASH_PIX_VERSION = 40960
EXIF_TAG_COLOR_SPACE = 40961
EXIF_TAG_PIXEL_X_DIMENSION = 40962
EXIF_TAG_PIXEL_Y_DIMENSION = 40963
EXIF_TAG_RELATED_SOUND_FILE = 40964
EXIF_TAG_INTEROPERABILITY_IFD_POINTER = 40965
EXIF_TAG_FLASH_ENERGY = 41483
EXIF_TAG_SPATIAL_FREQUENCY_RESPONSE = 41484
EXIF_TAG_FOCAL_PLANE_X_RESOLUTION = 41486
EXIF_TAG_FOCAL_PLANE_Y_RESOLUTION = 41487
EXIF_TAG_FOCAL_PLANE_RESOLUTION_UNIT = 41488
EXIF_TAG_SUBJECT_LOCATION = 41492
EXIF_TAG_EXPOSURE_INDEX = 41493
EXIF_TAG_SENSING_METHOD = 41495
EXIF_TAG_FILE_SOURCE = 41728
EXIF_TAG_SCENE_TYPE = 41729
EXIF_TAG_NEW_CFA_PATTERN = 41730
EXIF_TAG_CUSTOM_RENDERED = 41985
EXIF_TAG_EXPOSURE_MODE = 41986
EXIF_TAG_WHITE_BALANCE = 41987
EXIF_TAG_DIGITAL_ZOOM_RATIO = 41988
EXIF_TAG_FOCAL_LENGTH_IN_35MM_FILM = 41989
EXIF_TAG_SCENE_CAPTURE_TYPE = 41990
EXIF_TAG_GAIN_CONTROL = 41991
EXIF_TAG_CONTRAST = 41992
EXIF_TAG_SATURATION = 41993
EXIF_TAG_SHARPNESS = 41994
EXIF_TAG_DEVICE_SETTING_DESCRIPTION = 41995
EXIF_TAG_SUBJECT_DISTANCE_RANGE = 41996
EXIF_TAG_IMAGE_UNIQUE_ID = 42016
EXIF_TAG_CAMERA_OWNER_NAME = 42032
EXIF_TAG_BODY_SERIAL_NUMBER = 42033
EXIF_TAG_LENS_SPECIFICATION = 42034
EXIF_TAG_LENS_MAKE = 42035
EXIF_TAG_LENS_MODEL = 42036
EXIF_TAG_LENS_SERIAL_NUMBER = 42037
EXIF_TAG_COMPOSITE_IMAGE = 42080
EXIF_TAG_SOURCE_IMAGE_NUMBER_OF_COMPOSITE_IMAGE = 42081
EXIF_TAG_SOURCE_EXPOSURE_TIMES_OF_COMPOSITE_IMAGE = 42082
EXIF_TAG_GAMMA = 42240
EXIF_TAG_PRINT_IMAGE_MATCHING = 50341
EXIF_TAG_PADDING = 59932
end
@enum ExifFormat::UInt32 begin
EXIF_FORMAT_BYTE = 1
EXIF_FORMAT_ASCII = 2
EXIF_FORMAT_SHORT = 3
EXIF_FORMAT_LONG = 4
EXIF_FORMAT_RATIONAL = 5
EXIF_FORMAT_SBYTE = 6
EXIF_FORMAT_UNDEFINED = 7
EXIF_FORMAT_SSHORT = 8
EXIF_FORMAT_SLONG = 9
EXIF_FORMAT_SRATIONAL = 10
EXIF_FORMAT_FLOAT = 11
EXIF_FORMAT_DOUBLE = 12
end
mutable struct _ExifEntryPrivate end
ExifEntryPrivate = _ExifEntryPrivate
mutable struct _ExifEntry
tag::ExifTag
format::ExifFormat
components::Culong
data::Ptr{Cuchar}
size::Cuint
parent::Ptr{ExifContent}
priv::Ptr{ExifEntryPrivate}
end
function Base.getproperty(x::Ptr{_ExifEntry}, f::Symbol)
f === :tag && return Ptr{ExifTag}(x + 0)
f === :format && return Ptr{ExifFormat}(x + 4)
f === :components && return Ptr{Culong}(x + 8)
f === :data && return Ptr{Ptr{Cuchar}}(x + 16)
f === :size && return Ptr{Cuint}(x + 24)
f === :parent && return Ptr{Ptr{ExifContent}}(x + 32)
f === :priv && return Ptr{Ptr{ExifEntryPrivate}}(x + 40)
return getfield(x, f)
end
function Base.setproperty!(x::Ptr{_ExifEntry}, f::Symbol, v)
unsafe_store!(getproperty(x, f), v)
end
ExifEntry = _ExifEntry
function exif_content_get_entry(content, tag)
ccall((:exif_content_get_entry, libexif), Ptr{ExifEntry}, (Ptr{ExifContent}, ExifTag), content, tag)
end
function exif_entry_get_value(entry, val, maxlen)
ccall((:exif_entry_get_value, libexif), Ptr{Cchar}, (Ptr{ExifEntry}, Ptr{Cchar}, Cuint), entry, val, maxlen)
end
function exif_ifd_get_name(ifd)
ccall((:exif_ifd_get_name, libexif), Ptr{Cchar}, (ExifIfd,), ifd)
end
@enum ExifDataType::UInt32 begin
EXIF_DATA_TYPE_UNCOMPRESSED_CHUNKY = 0
EXIF_DATA_TYPE_UNCOMPRESSED_PLANAR = 1
EXIF_DATA_TYPE_UNCOMPRESSED_YCC = 2
EXIF_DATA_TYPE_COMPRESSED = 3
EXIF_DATA_TYPE_COUNT = 4
# EXIF_DATA_TYPE_UNKNOWN = 4
end
@enum ExifSupportLevel::UInt32 begin
EXIF_SUPPORT_LEVEL_UNKNOWN = 0
EXIF_SUPPORT_LEVEL_NOT_RECORDED = 1
EXIF_SUPPORT_LEVEL_MANDATORY = 2
EXIF_SUPPORT_LEVEL_OPTIONAL = 3
end
function exif_tag_from_name(name)
ccall((:exif_tag_from_name, libexif), ExifTag, (Ptr{Cchar},), name)
end
function exif_tag_get_name_in_ifd(tag, ifd)
ccall((:exif_tag_get_name_in_ifd, libexif), Ptr{Cchar}, (ExifTag, ExifIfd), tag, ifd)
end
function exif_tag_get_title_in_ifd(tag, ifd)
ccall((:exif_tag_get_title_in_ifd, libexif), Ptr{Cchar}, (ExifTag, ExifIfd), tag, ifd)
end
function exif_tag_get_description_in_ifd(tag, ifd)
ccall((:exif_tag_get_description_in_ifd, libexif), Ptr{Cchar}, (ExifTag, ExifIfd), tag, ifd)
end
function exif_tag_get_support_level_in_ifd(tag, ifd, t)
ccall((:exif_tag_get_support_level_in_ifd, libexif), ExifSupportLevel, (ExifTag, ExifIfd, ExifDataType), tag, ifd, t)
end
function exif_tag_get_name(tag)
ccall((:exif_tag_get_name, libexif), Ptr{Cchar}, (ExifTag,), tag)
end
function exif_tag_get_title(tag)
ccall((:exif_tag_get_title, libexif), Ptr{Cchar}, (ExifTag,), tag)
end
function exif_tag_get_description(tag)
ccall((:exif_tag_get_description, libexif), Ptr{Cchar}, (ExifTag,), tag)
end
function exif_tag_table_get_tag(n)
ccall((:exif_tag_table_get_tag, libexif), ExifTag, (Cuint,), n)
end
function exif_tag_table_get_name(n)
ccall((:exif_tag_table_get_name, libexif), Ptr{Cchar}, (Cuint,), n)
end
function exif_tag_table_count()
ccall((:exif_tag_table_count, libexif), Cuint, ())
end
function exif_format_get_name(format)
ccall((:exif_format_get_name, libexif), Ptr{Cchar}, (ExifFormat,), format)
end
function exif_format_get_size(format)
ccall((:exif_format_get_size, libexif), Cuchar, (ExifFormat,), format)
end
ExifByte = Cuchar
ExifSByte = Int8
ExifAscii = Ptr{Cchar}
ExifShort = UInt16
ExifSShort = Int16
ExifLong = UInt32
ExifSLong = Int32
struct ExifRational
numerator::ExifLong
denominator::ExifLong
end
ExifUndefined = Cchar
struct ExifSRational
numerator::ExifSLong
denominator::ExifSLong
end
function exif_get_short(b, order)
ccall((:exif_get_short, libexif), ExifShort, (Ptr{Cuchar}, ExifByteOrder), b, order)
end
function exif_get_sshort(b, order)
ccall((:exif_get_sshort, libexif), ExifSShort, (Ptr{Cuchar}, ExifByteOrder), b, order)
end
function exif_get_long(b, order)
ccall((:exif_get_long, libexif), ExifLong, (Ptr{Cuchar}, ExifByteOrder), b, order)
end
function exif_get_slong(b, order)
ccall((:exif_get_slong, libexif), ExifSLong, (Ptr{Cuchar}, ExifByteOrder), b, order)
end
function exif_get_rational(b, order)
ccall((:exif_get_rational, libexif), ExifRational, (Ptr{Cuchar}, ExifByteOrder), b, order)
end
function exif_get_srational(b, order)
ccall((:exif_get_srational, libexif), ExifSRational, (Ptr{Cuchar}, ExifByteOrder), b, order)
end
function exif_set_short(b, order, value)
ccall((:exif_set_short, libexif), Cvoid, (Ptr{Cuchar}, ExifByteOrder, ExifShort), b, order, value)
end
function exif_set_sshort(b, order, value)
ccall((:exif_set_sshort, libexif), Cvoid, (Ptr{Cuchar}, ExifByteOrder, ExifSShort), b, order, value)
end
function exif_set_long(b, order, value)
ccall((:exif_set_long, libexif), Cvoid, (Ptr{Cuchar}, ExifByteOrder, ExifLong), b, order, value)
end
function exif_set_slong(b, order, value)
ccall((:exif_set_slong, libexif), Cvoid, (Ptr{Cuchar}, ExifByteOrder, ExifSLong), b, order, value)
end
function exif_set_rational(b, order, value)
ccall((:exif_set_rational, libexif), Cvoid, (Ptr{Cuchar}, ExifByteOrder, ExifRational), b, order, value)
end
function exif_set_srational(b, order, value)
ccall((:exif_set_srational, libexif), Cvoid, (Ptr{Cuchar}, ExifByteOrder, ExifSRational), b, order, value)
end
function exif_convert_utf16_to_utf8(out, in, maxlen)
ccall((:exif_convert_utf16_to_utf8, libexif), Cvoid, (Ptr{Cchar}, Ptr{Cuchar}, Cint), out, in, maxlen)
end
function exif_array_set_byte_order(arg1, arg2, arg3, o_orig, o_new)
ccall((:exif_array_set_byte_order, libexif), Cvoid, (ExifFormat, Ptr{Cuchar}, Cuint, ExifByteOrder, ExifByteOrder), arg1, arg2, arg3, o_orig, o_new)
end
# typedef void * ( * ExifMemAllocFunc ) ( ExifLong s )
ExifMemAllocFunc = Ptr{Cvoid}
# typedef void * ( * ExifMemReallocFunc ) ( void * p , ExifLong s )
ExifMemReallocFunc = Ptr{Cvoid}
# typedef void ( * ExifMemFreeFunc ) ( void * p )
ExifMemFreeFunc = Ptr{Cvoid}
mutable struct _ExifMem end
ExifMem = _ExifMem
function exif_mem_new(a, r, f)
ccall((:exif_mem_new, libexif), Ptr{ExifMem}, (ExifMemAllocFunc, ExifMemReallocFunc, ExifMemFreeFunc), a, r, f)
end
function exif_mem_ref(arg1)
ccall((:exif_mem_ref, libexif), Cvoid, (Ptr{ExifMem},), arg1)
end
function exif_mem_unref(arg1)
ccall((:exif_mem_unref, libexif), Cvoid, (Ptr{ExifMem},), arg1)
end
function exif_mem_alloc(m, s)
ccall((:exif_mem_alloc, libexif), Ptr{UInt8}, (Ptr{ExifMem}, ExifLong), m, s)
end
function exif_mem_realloc(m, p, s)
ccall((:exif_mem_realloc, libexif), Ptr{Cvoid}, (Ptr{ExifMem}, Ptr{Cvoid}, ExifLong), m, p, s)
end
function exif_mem_free(m, p)
ccall((:exif_mem_free, libexif), Cvoid, (Ptr{ExifMem}, Ptr{Cvoid}), m, p)
end
function exif_mem_new_default()
ccall((:exif_mem_new_default, libexif), Ptr{ExifMem}, ())
end
function exif_entry_new()
ccall((:exif_entry_new, libexif), Ptr{ExifEntry}, ())
end
function exif_entry_new_mem(arg1)
ccall((:exif_entry_new_mem, libexif), Ptr{ExifEntry}, (Ptr{ExifMem},), arg1)
end
function exif_entry_ref(entry)
ccall((:exif_entry_ref, libexif), Cvoid, (Ptr{ExifEntry},), entry)
end
function exif_entry_unref(entry)
ccall((:exif_entry_unref, libexif), Cvoid, (Ptr{ExifEntry},), entry)
end
function exif_entry_free(entry)
ccall((:exif_entry_free, libexif), Cvoid, (Ptr{ExifEntry},), entry)
end
function exif_entry_initialize(e, tag)
ccall((:exif_entry_initialize, libexif), Cvoid, (Ptr{ExifEntry}, ExifTag), e, tag)
end
function exif_entry_fix(entry)
ccall((:exif_entry_fix, libexif), Cvoid, (Ptr{ExifEntry},), entry)
end
function exif_entry_dump(entry, indent)
ccall((:exif_entry_dump, libexif), Cvoid, (Ptr{ExifEntry}, Cuint), entry, indent)
end
mutable struct _ExifLog end
ExifLog = _ExifLog
function exif_log_new()
ccall((:exif_log_new, libexif), Ptr{ExifLog}, ())
end
function exif_log_new_mem(arg1)
ccall((:exif_log_new_mem, libexif), Ptr{ExifLog}, (Ptr{ExifMem},), arg1)
end
function exif_log_ref(log)
ccall((:exif_log_ref, libexif), Cvoid, (Ptr{ExifLog},), log)
end
function exif_log_unref(log)
ccall((:exif_log_unref, libexif), Cvoid, (Ptr{ExifLog},), log)
end
function exif_log_free(log)
ccall((:exif_log_free, libexif), Cvoid, (Ptr{ExifLog},), log)
end
@enum ExifLogCode::UInt32 begin
EXIF_LOG_CODE_NONE = 0
EXIF_LOG_CODE_DEBUG = 1
EXIF_LOG_CODE_NO_MEMORY = 2
EXIF_LOG_CODE_CORRUPT_DATA = 3
end
function exif_log_code_get_title(code)
ccall((:exif_log_code_get_title, libexif), Ptr{Cchar}, (ExifLogCode,), code)
end
function exif_log_code_get_message(code)
ccall((:exif_log_code_get_message, libexif), Ptr{Cchar}, (ExifLogCode,), code)
end
# typedef void ( * ExifLogFunc ) ( ExifLog * log , ExifLogCode , char * domain , char * format , va_list args , void * data )
ExifLogFunc = Ptr{Cvoid}
function exif_log_set_func(log, func, data)
ccall((:exif_log_set_func, libexif), Cvoid, (Ptr{ExifLog}, ExifLogFunc, Ptr{Cvoid}), log, func, data)
end
mutable struct _ExifDataPrivate end
ExifDataPrivate = _ExifDataPrivate
struct _ExifData
ifd::NTuple{5, Ptr{ExifContent}}
data::Ptr{Cuchar}
size::Cuint
priv::Ptr{ExifDataPrivate}
end
ExifData = _ExifData
mutable struct _ExifMnoteData end
ExifMnoteData = _ExifMnoteData
function exif_mnote_data_ref(arg1)
ccall((:exif_mnote_data_ref, libexif), Cvoid, (Ptr{ExifMnoteData},), arg1)
end
function exif_mnote_data_unref(arg1)
ccall((:exif_mnote_data_unref, libexif), Cvoid, (Ptr{ExifMnoteData},), arg1)
end
function exif_mnote_data_load(d, buf, buf_size)
ccall((:exif_mnote_data_load, libexif), Cvoid, (Ptr{ExifMnoteData}, Ptr{Cuchar}, Cuint), d, buf, buf_size)
end
function exif_mnote_data_save(d, buf, buf_size)
ccall((:exif_mnote_data_save, libexif), Cvoid, (Ptr{ExifMnoteData}, Ptr{Ptr{Cuchar}}, Ptr{Cuint}), d, buf, buf_size)
end
function exif_mnote_data_count(d)
ccall((:exif_mnote_data_count, libexif), Cuint, (Ptr{ExifMnoteData},), d)
end
function exif_mnote_data_get_id(d, n)
ccall((:exif_mnote_data_get_id, libexif), Cuint, (Ptr{ExifMnoteData}, Cuint), d, n)
end
function exif_mnote_data_get_name(d, n)
ccall((:exif_mnote_data_get_name, libexif), Ptr{Cchar}, (Ptr{ExifMnoteData}, Cuint), d, n)
end
function exif_mnote_data_get_title(d, n)
ccall((:exif_mnote_data_get_title, libexif), Ptr{Cchar}, (Ptr{ExifMnoteData}, Cuint), d, n)
end
function exif_mnote_data_get_description(d, n)
ccall((:exif_mnote_data_get_description, libexif), Ptr{Cchar}, (Ptr{ExifMnoteData}, Cuint), d, n)
end
function exif_mnote_data_get_value(d, n, val, maxlen)
ccall((:exif_mnote_data_get_value, libexif), Ptr{Cchar}, (Ptr{ExifMnoteData}, Cuint, Ptr{Cchar}, Cuint), d, n, val, maxlen)
end
function exif_mnote_data_log(arg1, arg2)
ccall((:exif_mnote_data_log, libexif), Cvoid, (Ptr{ExifMnoteData}, Ptr{ExifLog}), arg1, arg2)
end
function exif_data_new()
ccall((:exif_data_new, libexif), Ptr{ExifData}, ())
end
function exif_data_new_mem(arg1)
ccall((:exif_data_new_mem, libexif), Ptr{ExifData}, (Ptr{ExifMem},), arg1)
end
function exif_data_new_from_file(path)
ccall((:exif_data_new_from_file, libexif), Ptr{ExifData}, (Ptr{Cchar},), path)
end
function exif_data_new_from_data(data, size)
ccall((:exif_data_new_from_data, libexif), Ptr{ExifData}, (Ptr{Cuchar}, Cuint), data, size)
end
function exif_data_load_data(data, d, size)
ccall((:exif_data_load_data, libexif), Cvoid, (Ptr{ExifData}, Ptr{Cuchar}, Cuint), data, d, size)
end
function exif_data_save_data(data, d, ds)
ccall((:exif_data_save_data, libexif), Cvoid, (Ptr{ExifData}, Ptr{Ptr{Cuchar}}, Ptr{Cuint}), data, d, ds)
end
function exif_data_ref(data)
ccall((:exif_data_ref, libexif), Cvoid, (Ptr{ExifData},), data)
end
function exif_data_unref(data)
ccall((:exif_data_unref, libexif), Cvoid, (Ptr{ExifData},), data)
end
function exif_data_free(data)
ccall((:exif_data_free, libexif), Cvoid, (Ptr{ExifData},), data)
end
function exif_data_get_byte_order(data)
ccall((:exif_data_get_byte_order, libexif), ExifByteOrder, (Ptr{ExifData},), data)
end
function exif_data_set_byte_order(data, order)
ccall((:exif_data_set_byte_order, libexif), Cvoid, (Ptr{ExifData}, ExifByteOrder), data, order)
end
function exif_data_get_mnote_data(d)
ccall((:exif_data_get_mnote_data, libexif), Ptr{ExifMnoteData}, (Ptr{ExifData},), d)
end
function exif_data_fix(d)
ccall((:exif_data_fix, libexif), Cvoid, (Ptr{ExifData},), d)
end
# typedef void ( * ExifDataForeachContentFunc ) ( ExifContent * , void * user_data )
ExifDataForeachContentFunc = Ptr{Cvoid}
function exif_data_foreach_content(data, func, user_data)
ccall((:exif_data_foreach_content, libexif), Cvoid, (Ptr{ExifData}, ExifDataForeachContentFunc, Ptr{Cvoid}), data, func, user_data)
end
@enum ExifDataOption::UInt32 begin
EXIF_DATA_OPTION_IGNORE_UNKNOWN_TAGS = 1
EXIF_DATA_OPTION_FOLLOW_SPECIFICATION = 2
EXIF_DATA_OPTION_DONT_CHANGE_MAKER_NOTE = 4
end
function exif_data_option_get_name(o)
ccall((:exif_data_option_get_name, libexif), Ptr{Cchar}, (ExifDataOption,), o)
end
function exif_data_option_get_description(o)
ccall((:exif_data_option_get_description, libexif), Ptr{Cchar}, (ExifDataOption,), o)
end
function exif_data_set_option(d, o)
ccall((:exif_data_set_option, libexif), Cvoid, (Ptr{ExifData}, ExifDataOption), d, o)
end
function exif_data_unset_option(d, o)
ccall((:exif_data_unset_option, libexif), Cvoid, (Ptr{ExifData}, ExifDataOption), d, o)
end
function exif_data_set_data_type(d, dt)
ccall((:exif_data_set_data_type, libexif), Cvoid, (Ptr{ExifData}, ExifDataType), d, dt)
end
function exif_data_get_data_type(d)
ccall((:exif_data_get_data_type, libexif), ExifDataType, (Ptr{ExifData},), d)
end
function exif_data_dump(data)
ccall((:exif_data_dump, libexif), Cvoid, (Ptr{ExifData},), data)
end
function exif_data_log(data, log)
ccall((:exif_data_log, libexif), Cvoid, (Ptr{ExifData}, Ptr{ExifLog}), data, log)
end
function exif_content_new()
ccall((:exif_content_new, libexif), Ptr{ExifContent}, ())
end
function exif_content_new_mem(arg1)
ccall((:exif_content_new_mem, libexif), Ptr{ExifContent}, (Ptr{ExifMem},), arg1)
end
function exif_content_ref(content)
ccall((:exif_content_ref, libexif), Cvoid, (Ptr{ExifContent},), content)
end
function exif_content_unref(content)
ccall((:exif_content_unref, libexif), Cvoid, (Ptr{ExifContent},), content)
end
function exif_content_free(content)
ccall((:exif_content_free, libexif), Cvoid, (Ptr{ExifContent},), content)
end
function exif_content_add_entry(c, entry)
ccall((:exif_content_add_entry, libexif), Cvoid, (Ptr{ExifContent}, Ptr{ExifEntry}), c, entry)
end
function exif_content_remove_entry(c, e)
ccall((:exif_content_remove_entry, libexif), Cvoid, (Ptr{ExifContent}, Ptr{ExifEntry}), c, e)
end
function exif_content_fix(c)
ccall((:exif_content_fix, libexif), Cvoid, (Ptr{ExifContent},), c)
end
# typedef void ( * ExifContentForeachEntryFunc ) ( ExifEntry * , void * user_data )
ExifContentForeachEntryFunc = Ptr{Cvoid}
function exif_content_foreach_entry(content, func, user_data)
ccall((:exif_content_foreach_entry, libexif), Cvoid, (Ptr{ExifContent}, ExifContentForeachEntryFunc, Ptr{Cvoid}), content, func, user_data)
end
function exif_content_dump(content, indent)
ccall((:exif_content_dump, libexif), Cvoid, (Ptr{ExifContent}, Cuint), content, indent)
end
function exif_content_log(content, log)
ccall((:exif_content_log, libexif), Cvoid, (Ptr{ExifContent}, Ptr{ExifLog}), content, log)
end
mutable struct _ExifLoader end
ExifLoader = _ExifLoader
function exif_loader_new()
ccall((:exif_loader_new, libexif), Ptr{ExifLoader}, ())
end
function exif_loader_new_mem(mem)
ccall((:exif_loader_new_mem, libexif), Ptr{ExifLoader}, (Ptr{ExifMem},), mem)
end
function exif_loader_ref(loader)
ccall((:exif_loader_ref, libexif), Cvoid, (Ptr{ExifLoader},), loader)
end
function exif_loader_unref(loader)
ccall((:exif_loader_unref, libexif), Cvoid, (Ptr{ExifLoader},), loader)
end
function exif_loader_write_file(loader, fname)
ccall((:exif_loader_write_file, libexif), Cvoid, (Ptr{ExifLoader}, Ptr{Cchar}), loader, fname)
end
function exif_loader_write(loader, buf, sz)
ccall((:exif_loader_write, libexif), Cuchar, (Ptr{ExifLoader}, Ptr{Cuchar}, Cuint), loader, buf, sz)
end
function exif_loader_reset(loader)
ccall((:exif_loader_reset, libexif), Cvoid, (Ptr{ExifLoader},), loader)
end
function exif_loader_get_data(loader)
ccall((:exif_loader_get_data, libexif), Ptr{ExifData}, (Ptr{ExifLoader},), loader)
end
function exif_loader_get_buf(loader, buf, buf_size)
ccall((:exif_loader_get_buf, libexif), Cvoid, (Ptr{ExifLoader}, Ptr{Ptr{Cuchar}}, Ptr{Cuint}), loader, buf, buf_size)
end
function exif_loader_log(loader, log)
ccall((:exif_loader_log, libexif), Cvoid, (Ptr{ExifLoader}, Ptr{ExifLog}), loader, log)
end
@enum ExifTagGPS::UInt16 begin
EXIF_TAG_GPS_VERSION_ID = 0x0000
EXIF_TAG_GPS_LATITUDE_REF = 0x0001
EXIF_TAG_GPS_LATITUDE = 0x0002
EXIF_TAG_GPS_LONGITUDE_REF = 0x0003
EXIF_TAG_GPS_LONGITUDE = 0x0004
EXIF_TAG_GPS_ALTITUDE_REF = 0x0005
EXIF_TAG_GPS_ALTITUDE = 0x0006
EXIF_TAG_GPS_TIME_STAMP = 0x0007
EXIF_TAG_GPS_SATELLITES = 0x0008
EXIF_TAG_GPS_STATUS = 0x0009
EXIF_TAG_GPS_MEASURE_MODE = 0x000a
EXIF_TAG_GPS_DOP = 0x000b
EXIF_TAG_GPS_SPEED_REF = 0x000c
EXIF_TAG_GPS_SPEED = 0x000d
EXIF_TAG_GPS_TRACK_REF = 0x000e
EXIF_TAG_GPS_TRACK = 0x000f
EXIF_TAG_GPS_IMG_DIRECTION_REF = 0x0010
EXIF_TAG_GPS_IMG_DIRECTION = 0x0011
EXIF_TAG_GPS_MAP_DATUM = 0x0012
EXIF_TAG_GPS_DEST_LATITUDE_REF = 0x0013
EXIF_TAG_GPS_DEST_LATITUDE = 0x0014
EXIF_TAG_GPS_DEST_LONGITUDE_REF = 0x0015
EXIF_TAG_GPS_DEST_LONGITUDE = 0x0016
EXIF_TAG_GPS_DEST_BEARING_REF = 0x0017
EXIF_TAG_GPS_DEST_BEARING = 0x0018
EXIF_TAG_GPS_DEST_DISTANCE_REF = 0x0019
EXIF_TAG_GPS_DEST_DISTANCE = 0x001a
EXIF_TAG_GPS_PROCESSING_METHOD = 0x001b
EXIF_TAG_GPS_AREA_INFORMATION = 0x001c
EXIF_TAG_GPS_DATE_STAMP = 0x001d
EXIF_TAG_GPS_DIFFERENTIAL = 0x001e
EXIF_TAG_GPS_H_POSITIONING_ERROR = 0x001f
# EXIF_TAG_UNKNOWN_C4A5 = EXIF_TAG_PRINT_IMAGE_MATCHING
EXIF_TAG_UNKNOWN_C4A5 = 0xc4a5
# EXIF_TAG_SUBSEC_TIME = EXIF_TAG_SUB_SEC_TIME
EXIF_TAG_SUBSEC_TIME = 0x9290
end
end # module
| ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | code | 218 | module ExifViewer
include("../lib/LibExif.jl")
using .LibExif
using ColorTypes
using JpegTurbo
include("utils.jl")
include("read.jl")
include("write.jl")
include("precompile.jl")
export read_tags, write_tags
end
| ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | code | 5852 | using SnoopPrecompile
# setup for precompilation
@precompile_setup begin
file = UInt8[0xff, 0xd8, 0xff, 0xe1, 0x00, 0xa5, 0x45, 0x78, 0x69, 0x66, 0x00, 0x00, 0x49, 0x49, 0x2a, 0x00, 0x08, 0x00, 0x00, 0x00, 0x06, 0x00, 0x1a,
0x01, 0x05, 0x00, 0x01, 0x00, 0x00, 0x00, 0x56, 0x00, 0x00, 0x00, 0x1b, 0x01, 0x05, 0x00, 0x01, 0x00, 0x00, 0x00, 0x5e, 0x00, 0x00, 0x00, 0x28, 0x01, 0x03, 0x00, 0x01, 0x00, 0x00, 0x00, 0x02, 0x00, 0x00, 0x00, 0x3b, 0x01, 0x02, 0x00, 0x07, 0x00, 0x00, 0x00, 0x66, 0x00, 0x00, 0x00, 0x13, 0x02, 0x03, 0x00, 0x01, 0x00, 0x00, 0x00, 0x01, 0x00, 0x00, 0x00, 0x69, 0x87, 0x04, 0x00, 0x01, 0x00, 0x00, 0x00, 0x6e, 0x00, 0x00, 0x00, 0x00, 0x00, 0x00, 0x00, 0x48, 0x00, 0x00, 0x00, 0x01, 0x00, 0x00, 0x00, 0x48, 0x00, 0x00, 0x00, 0x01, 0x00, 0x00, 0x00, 0x41, 0x73, 0x68, 0x77, 0x61, 0x6e, 0x69, 0x00, 0x06, 0x00, 0x00, 0x90, 0x07, 0x00, 0x04, 0x00, 0x00, 0x00, 0x30, 0x32, 0x31, 0x30, 0x01, 0x91, 0x07, 0x00, 0x04, 0x00, 0x00, 0x00, 0x01, 0x02, 0x03, 0x00, 0x00, 0xa0, 0x07, 0x00, 0x04, 0x00, 0x00, 0x00, 0x30, 0x31, 0x30, 0x30, 0x01, 0xa0, 0x03, 0x00, 0x01, 0x00, 0x00, 0x00, 0xff, 0xff, 0xe0, 0x00, 0x10, 0x4a, 0x46, 0x49, 0x46, 0x00, 0x01, 0x01, 0x00, 0x00, 0x01, 0x00, 0x01, 0x00,
0x00, 0xff, 0xdb, 0x00, 0x43, 0x00, 0x03, 0x02, 0x02, 0x02, 0x02, 0x02, 0x03, 0x02, 0x02, 0x02, 0x03, 0x03, 0x03, 0x03, 0x04, 0x06, 0x04, 0x04, 0x04, 0x04, 0x04, 0x08, 0x06, 0x06, 0x05, 0x06, 0x09, 0x08, 0x0a, 0x0a, 0x09, 0x08, 0x09, 0x09, 0x0a, 0x0c, 0x0f, 0x0c, 0x0a, 0x0b, 0x0e, 0x0b, 0x09, 0x09, 0x0d, 0x11, 0x0d, 0x0e, 0x0f, 0x10, 0x10, 0x11, 0x10, 0x0a, 0x0c, 0x12, 0x13, 0x12, 0x10, 0x13, 0x0f, 0x10, 0x10, 0x10, 0xff, 0xdb, 0x00, 0x43, 0x01, 0x03, 0x03, 0x03, 0x04, 0x03, 0x04, 0x08, 0x04, 0x04, 0x08, 0x10, 0x0b, 0x09, 0x0b, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0x10, 0xff, 0xc0, 0x00, 0x11, 0x08, 0x00, 0x01, 0x00, 0x01, 0x03, 0x01, 0x22, 0x00, 0x02, 0x11, 0x01, 0x03, 0x11, 0x01, 0xff, 0xc4, 0x00, 0x1f, 0x00,
0x00, 0x01, 0x05, 0x01, 0x01, 0x01, 0x01, 0x01, 0x01, 0x00, 0x00, 0x00, 0x00, 0x00, 0x00, 0x00, 0x00, 0x01, 0x02, 0x03, 0x04, 0x05, 0x06, 0x07, 0x08, 0x09, 0x0a, 0x0b, 0xff, 0xc4, 0x00, 0xb5, 0x10, 0x00, 0x02, 0x01, 0x03, 0x03, 0x02, 0x04, 0x03, 0x05, 0x05, 0x04, 0x04, 0x00, 0x00, 0x01, 0x7d, 0x01, 0x02, 0x03, 0x00, 0x04, 0x11, 0x05, 0x12, 0x21, 0x31, 0x41, 0x06, 0x13, 0x51, 0x61, 0x07, 0x22, 0x71, 0x14, 0x32, 0x81, 0x91, 0xa1, 0x08, 0x23, 0x42, 0xb1, 0xc1, 0x15, 0x52, 0xd1, 0xf0, 0x24, 0x33, 0x62, 0x72, 0x82, 0x09, 0x0a, 0x16, 0x17, 0x18, 0x19, 0x1a, 0x25, 0x26, 0x27, 0x28, 0x29, 0x2a, 0x34, 0x35, 0x36, 0x37, 0x38, 0x39, 0x3a, 0x43, 0x44, 0x45, 0x46, 0x47, 0x48, 0x49, 0x4a, 0x53, 0x54, 0x55, 0x56, 0x57, 0x58, 0x59, 0x5a, 0x63, 0x64, 0x65, 0x66, 0x67, 0x68, 0x69, 0x6a, 0x73, 0x74, 0x75, 0x76, 0x77, 0x78, 0x79, 0x7a, 0x83, 0x84, 0x85, 0x86, 0x87, 0x88, 0x89, 0x8a, 0x92, 0x93, 0x94, 0x95, 0x96, 0x97, 0x98, 0x99, 0x9a, 0xa2, 0xa3, 0xa4, 0xa5, 0xa6, 0xa7, 0xa8, 0xa9,
0xaa, 0xb2, 0xb3, 0xb4, 0xb5, 0xb6, 0xb7, 0xb8, 0xb9, 0xba, 0xc2, 0xc3, 0xc4, 0xc5, 0xc6, 0xc7, 0xc8, 0xc9, 0xca, 0xd2, 0xd3, 0xd4, 0xd5, 0xd6, 0xd7, 0xd8, 0xd9, 0xda, 0xe1, 0xe2, 0xe3, 0xe4, 0xe5, 0xe6, 0xe7, 0xe8, 0xe9, 0xea, 0xf1, 0xf2, 0xf3, 0xf4, 0xf5, 0xf6, 0xf7, 0xf8, 0xf9, 0xfa, 0xff, 0xc4, 0x00, 0x1f, 0x01, 0x00, 0x03, 0x01, 0x01, 0x01, 0x01, 0x01, 0x01, 0x01, 0x01, 0x01, 0x00, 0x00, 0x00, 0x00, 0x00, 0x00, 0x01, 0x02, 0x03, 0x04, 0x05, 0x06, 0x07, 0x08, 0x09, 0x0a, 0x0b, 0xff, 0xc4, 0x00, 0xb5, 0x11, 0x00, 0x02, 0x01, 0x02, 0x04, 0x04, 0x03, 0x04, 0x07, 0x05, 0x04, 0x04, 0x00, 0x01, 0x02, 0x77, 0x00, 0x01, 0x02, 0x03, 0x11, 0x04, 0x05, 0x21, 0x31, 0x06, 0x12, 0x41, 0x51, 0x07, 0x61, 0x71, 0x13, 0x22, 0x32, 0x81, 0x08, 0x14, 0x42, 0x91, 0xa1, 0xb1, 0xc1, 0x09, 0x23, 0x33, 0x52, 0xf0, 0x15, 0x62, 0x72, 0xd1, 0x0a, 0x16, 0x24, 0x34, 0xe1, 0x25, 0xf1, 0x17, 0x18, 0x19, 0x1a, 0x26, 0x27, 0x28, 0x29, 0x2a, 0x35, 0x36, 0x37, 0x38, 0x39, 0x3a, 0x43, 0x44, 0x45,
0x46, 0x47, 0x48, 0x49, 0x4a, 0x53, 0x54, 0x55, 0x56, 0x57, 0x58, 0x59, 0x5a, 0x63, 0x64, 0x65, 0x66, 0x67, 0x68, 0x69, 0x6a, 0x73, 0x74, 0x75, 0x76, 0x77, 0x78, 0x79, 0x7a, 0x82, 0x83, 0x84, 0x85, 0x86, 0x87, 0x88, 0x89, 0x8a, 0x92, 0x93, 0x94, 0x95, 0x96, 0x97, 0x98, 0x99, 0x9a, 0xa2, 0xa3, 0xa4, 0xa5, 0xa6, 0xa7, 0xa8, 0xa9, 0xaa, 0xb2, 0xb3, 0xb4, 0xb5, 0xb6, 0xb7, 0xb8, 0xb9, 0xba, 0xc2, 0xc3, 0xc4, 0xc5, 0xc6, 0xc7, 0xc8, 0xc9, 0xca, 0xd2, 0xd3, 0xd4, 0xd5, 0xd6, 0xd7, 0xd8, 0xd9, 0xda, 0xe2, 0xe3, 0xe4, 0xe5, 0xe6, 0xe7, 0xe8, 0xe9, 0xea, 0xf2, 0xf3, 0xf4, 0xf5, 0xf6, 0xf7, 0xf8, 0xf9, 0xfa, 0xff, 0xda, 0x00, 0x0c, 0x03, 0x01, 0x00, 0x02, 0x11, 0x03, 0x11, 0x00, 0x3f, 0x00, 0xfc, 0xaa, 0xa2, 0x8a, 0x28, 0x03, 0xff, 0xd9]
path = joinpath(tempdir(), "tmp.jpg")
write(path, file)
tags = Dict{String, String}(
"EXIF_TAG_MAKE" => "test",
"EXIF_TAG_ORIENTATION" => "Top-left",
"EXIF_TAG_X_RESOLUTION" => "300",
"EXIF_TAG_Y_RESOLUTION" => "300",
)
imgs_list = Any[
rand(Gray{ColorTypes.N0f8}, 32, 32),
rand(RGB{ColorTypes.N0f8}, 32, 32),
rand(Gray{ColorTypes.Float64}, 32, 32),
rand(RGB{Float64}, 32, 32),
rand(ColorTypes.N0f8, 32, 32),
rand(ColorTypes.Float64, 32, 32)
]
# precompiling the calls for future, therefore future calls are faster
@precompile_all_calls begin
read_tags(file; read_all=true)
read_tags(path; read_all=true)
open(path, "r") do io
read_tags(io; read_all=true)
end
for img in imgs_list
write_tags(path; img, tags)
end
end
rm(path)
end | ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | code | 6230 |
const IFDS_ALL_FIELDS = 1:5 # Specifies all IFDs(Image File Directory)
"""
read_tags(data::Vector{UInt8}; kwargs...})
read_tags(filepath::AbstractString; kwargs...)
read_tags(io::IO; kwargs...)
Read EXIF tags from the input source data. Return an empty dictionary if the source data doesn't contain EXIF tags.
#### Keyword Arguments
- `ifds::Union{Int,NTuple,UnitRange}` : Defines which IFD(Image file directory) to search in for the EXIF tags. Default is all ifds i.e. 1:5.
- `read_all::Bool` : Defines if all EXIF tags are to be read or not. By default, `read_all` is true.
- `tags::Vector{LibExif.ExifTag}` : Defines which tags to search, in case `read_all` is false. When `read_all` is false, tags that need to be searched need to defined manually. Tags can be provided using bunch of methods but its suggested to supply a vector of strings with each string representing a EXIF tag i.e. ["`EXIF_TAG_FLASH_PIX_VERSION`", "`EXIF_TAG_ORIENTATION`"]
- `extract_thumbnail::Bool` : Defines whether to read the thumbnail data or not. By default, `extract_thumbnail` is false.
- `read_mnote::Bool` : Defines whether to read the mnote(MakerNote) tags data or not. By default, `read_mnote` is false.
List of all available tags to search is available here: https://libexif.github.io/internals/exif-tag_8h.html
#### Examples
```jl
julia> using TestImages, ExifViewer
julia> filepath = testimage("earth_apollo17.jpg", download_only=true)
julia> io = open(filepath, "r")
julia> read_tags(io; read_all=false, tags=["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"])
Dict{Any, Any} with 2 entries:
"EXIF_TAG_FLASH_PIX_VERSION" => "FlashPix Version 1.0"
"EXIF_TAG_ORIENTATION" => "Top-left"
julia> read_tags(filepath; read_all=false, tags=["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"])
Dict{Any, Any} with 2 entries:
"EXIF_TAG_FLASH_PIX_VERSION" => "FlashPix Version 1.0"
"EXIF_TAG_ORIENTATION" => "Top-left"
julia> data = read(filepath)
julia> read_tags(data, read_all=false, tags=["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"])
Dict{Any, Any} with 2 entries:
"EXIF_TAG_FLASH_PIX_VERSION" => "FlashPix Version 1.0"
"EXIF_TAG_ORIENTATION" => "Top-left"
```
"""
function read_tags(
data::Vector{UInt8};
ifds::Union{Int,NTuple,UnitRange} = IFDS_ALL_FIELDS,
read_all::Bool = true,
tags::Union{AbstractVector,Tuple} = Vector{LibExif.ExifTag}([]),
extract_thumbnail::Bool = false,
read_mnote::Bool = false,
)
ed_ptr = LibExif.exif_data_new_from_data(data, length(data))
if (ed_ptr == C_NULL)
return error("Unable to read EXIF data: invalid pointer")
end
tags = normalize_exif_flag(tags)
typeassert(tags, Vector{LibExif.ExifTag})
result = Dict{String,String}()
thumbnail_data = UInt8[]
try
ed = unsafe_load(ed_ptr)
ifds = collect(ifds)
# ifds = read_all ? collect(1:numifds(ed)) : collect(ifds)
checkbounds(Bool, collect(1:numifds(ed)), ifds) ||
throw(BoundsError(collect(1:numifds(ed)), ifds))
tags = read_all ? tags : Set(tags)
str = Vector{Cuchar}(undef, 1024)
for i in ifds
content_ptr = ed.ifd[i]
if (content_ptr == C_NULL)
return error("Unable to read IFD:", i)
end
data = unsafe_load(content_ptr)
if data.count == 0
continue
end
res = unsafe_wrap(Array, data.entries, data.count)
for j = 1:data.count
entry = unsafe_load(res[j])
condition = read_all ? read_all : entry.tag in tags
if condition
LibExif.exif_entry_get_value(Ref(entry), str, length(str))
tag = String(copy(str))[1:max(findfirst(iszero, str) - 1, 1)]
# @info entry.tag entry.format
if string(entry.tag) ∉ keys(result)
tagname = string(entry.tag)
# to update name if its gps ifd
if (i == 4 && UInt16(entry.tag) in keys(Base.Enums.namemap(LibExif.ExifTagGPS)))
tagname = string(LibExif.ExifTagGPS(UInt16(entry.tag)))
end
result[tagname] = strip(tag)
end
end
if read_all == false
delete!(tags, entry.tag)
if tags == Set()
break
end
end
end
end
# not sure we should include this
# if isempty(tags) != true
# @info "Non-Existent Tags:" tags
# end
if (read_mnote == true)
md_ptr = LibExif.exif_data_get_mnote_data(ed_ptr)
if (md_ptr == C_NULL)
return error("Unable to read MNOTE data")
end
LibExif.exif_mnote_data_ref(md_ptr)
LibExif.exif_mnote_data_unref(md_ptr)
c = LibExif.exif_mnote_data_count(md_ptr)
for i = 0:c-1
mnote = LibExif.exif_mnote_data_get_name(md_ptr, i)
if (mnote == C_NULL)
continue
end
data = unsafe_string(mnote)
name = uppercase(replace(data, " " => "_")) # preprocess
LibExif.exif_mnote_data_get_value(md_ptr, i, str, length(str))
tag = String(copy(str))[1:max(findfirst(iszero, str) - 1, 1)]
if name ∉ keys(result)
result["MNOTE_"*name] = tag
end
end
end
if (extract_thumbnail == true)
thumbnail_size = Int(ed.size)
thumbnail_data = unsafe_wrap(Array, ed.data, thumbnail_size)
end
finally
LibExif.exif_data_unref(ed_ptr)
end
if (extract_thumbnail) return result, thumbnail_data
else return result end
end
function read_tags(filepath::AbstractString; kwargs...)
open(filepath, "r") do io
read_tags(read(io); kwargs...)
end
end
function read_tags(io::IO; kwargs...)
try
read_tags(read(io); kwargs...)
finally
close(io)
end
end
| ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | code | 7292 | """
numifds(ed::LibExif._ExifData)
Returns the number of IFDs in the EXIF data.
"""
function numifds(ed::LibExif._ExifData)
return length(ed.ifd)
end
"""
numentriesinifd(data::LibExif._ExifContent)
Returns the number of entries in the IFD.
"""
function numentriesinifd(data::LibExif._ExifContent)
return Int(data.count)
end
"""
normalize_exif_flag(flags::Union{AbstractVector,Tuple}) = map(normalize_exif_flag, flags)
normalize_exif_flag(flag::AbstractString) = normalize_exif_flag(Symbol(flag))
normalize_exif_flag(flag::Symbol) = getfield(LibExif, flag)
normalize_exif_flag(flag::LibExif.ExifTag) = flag
normalize_exif_flag(flag::Int) = LibExif.ExifTag(flag)
Converts the input to `LibExif.ExifTag` type if possible.
"""
normalize_exif_flag(flags::Union{AbstractVector,Tuple}) = map(normalize_exif_flag, flags)
normalize_exif_flag(flag::AbstractString) = normalize_exif_flag(Symbol(flag))
normalize_exif_flag(flag::Symbol) = getfield(LibExif, flag)
normalize_exif_flag(flag::LibExif.ExifTag) = flag
normalize_exif_flag(flag::Int) = LibExif.ExifTag(flag)
TagsDict = Dict(
LibExif.EXIF_TAG_COMPRESSION => Dict{String,UInt16}(
"Uncompressed" => 1,
"LZW compression" => 5,
"JPEG compression" => 6,
"Deflate/ZIP compression" => 8,
"PackBits compression" => 32773,
"" => 0,
),
LibExif.EXIF_TAG_COLOR_SPACE => Dict{String,UInt16}(
"sRGB" => 1,
"Adobe RGB" => 2,
"Uncalibrated" => 0xff,
"" => 0,
),
LibExif.EXIF_TAG_SUBJECT_DISTANCE_RANGE => Dict{String,UInt16}(
"Unknown" => 0,
"Macro" => 1,
"Close view" => 2,
"Distant view" => 3,
"" => 0,
),
LibExif.EXIF_TAG_ORIENTATION => Dict{String,UInt64}(
"Top-left" => 1,
"Top-right" => 2,
"Bottom-right" => 3,
"Bottom-left" => 4,
"Left-top" => 5,
"Right-top" => 6,
"Right-bottom" => 7,
"Left-bottom" => 8,
"" => 0,
),
LibExif.EXIF_TAG_METERING_MODE => Dict{String,UInt64}(
"Average" => 1,
"Avg" => 1,
"Center-weighted average" => 2,
"Center-weight" => 2,
"Spot" => 3,
"Multi spot" => 4,
"Pattern" => 5,
"Partial" => 6,
"Other" => 255,
"" => 0,
),
LibExif.EXIF_TAG_SENSING_METHOD => Dict{String,UInt64}(
"Not defined" => 1,
"One-chip color area sensor" => 2,
"Two-chip color area sensor" => 3,
"Three-chip color area sensor" => 4,
"Color sequential area sensor" => 5,
"Trilinear sensor" => 6,
"Color sequential linear sensor" => 7,
"" => 0,
),
LibExif.EXIF_TAG_FLASH => Dict{String,UInt64}(
"Flash did not fire" => 0x0000,
"No flash" => 0x0000,
"Flash fired" => 0x0001,
"Flash" => 0x0001,
"Yes" => 0x0001,
"Strobe return light not detected" => 0x0005,
"Without strobe" => 0x0005,
"Strobe return light detected" => 0x0007,
"With strobe" => 0x0007,
"Flash did not fire" => 0x0007,
"Flash fired, compulsory flash mode" => 0x0009,
"Flash fired, compulsory flash mode, return light not detected" => 0x000d,
"Flash fired, compulsory flash mode, return light detected" => 0x000f,
"Flash did not fire, compulsory flash mode" => 0x0010,
"Flash did not fire, auto mode" => 0x0018,
"Flash fired, auto mode" => 0x0019,
"Flash fired, auto mode, return light not detected" => 0x001d,
"Flash fired, auto mode, return light detected" => 0x001f,
"No flash function" => 0x0020,
"Flash fired, red-eye reduction mode" => 0x0041,
"Flash fired, red-eye reduction mode, return light not detected" => 0x0045,
"Flash fired, red-eye reduction mode, return light detected" => 0x0047,
"Flash fired, compulsory flash mode, red-eye reduction mode" => 0x0049,
"Flash fired, compulsory flash mode, red-eye reduction mode, return light not detected" =>
0x004d,
"Flash fired, compulsory flash mode, red-eye reduction mode, return light detected" =>
0x004f,
"Flash did not fire, auto mode, red-eye reduction mode" => 0x0058,
"Flash fired, auto mode, red-eye reduction mode" => 0x0059,
"Flash fired, auto mode, return light not detected, red-eye reduction mode" =>
0x005d,
"Flash fired, auto mode, return light detected, red-eye reduction mode" =>
0x005f,
"" => 0,
),
LibExif.EXIF_TAG_YCBCR_POSITIONING => Dict{String,UInt64}("Centered" => 1, "Co-sited" => 2, "" => 0),
LibExif.EXIF_TAG_RESOLUTION_UNIT => Dict{String,UInt64}(
"Inch" => 2,
"in" => 2,
"Centimeter" => 3,
"cm" => 3,
"" => 0,
),
LibExif.EXIF_TAG_FOCAL_PLANE_RESOLUTION_UNIT => Dict{String,UInt64}(
"Inch" => 2,
"in" => 2,
"Centimeter" => 3,
"cm" => 3,
),
LibExif.EXIF_TAG_PLANAR_CONFIGURATION => Dict{String,UInt16}(
"Chunky format" => 0,
"Planar format" => 1,
),
LibExif.EXIF_TAG_PHOTOMETRIC_INTERPRETATION => Dict{String,UInt16}(
"Reversed mono" => 0,
"Normal mono" => 1,
"RGB" => 2,
"Palette" => 3,
"CMYK" => 5,
"YCbCr" => 6,
"CieLAB" => 8,
),
LibExif.EXIF_TAG_CUSTOM_RENDERED => Dict{String,UInt16}(
"Normal process" => 0,
"Custom process" => 1,
),
LibExif.EXIF_TAG_EXPOSURE_MODE => Dict{String,UInt16}(
"Auto exposure" => 0,
"Manual exposure" => 1,
"Auto Bracket" => 2,
),
LibExif.EXIF_TAG_WHITE_BALANCE => Dict{String,UInt16}(
"Auto white balance" => 0,
"Manual white balance" => 1,
),
LibExif.EXIF_TAG_SCENE_CAPTURE_TYPE => Dict{String,UInt16}(
"Standard" => 0,
"Landscape" => 1,
"Portrait" => 2,
"Night scene" => 3,
),
LibExif.EXIF_TAG_GAIN_CONTROL => Dict{String,UInt16}(
"Normal" => 0,
"Low gain up" => 1,
"High gain up" => 2,
"Low gain down" => 3,
"High gain down" => 4,
),
LibExif.EXIF_TAG_SATURATION => Dict{String,UInt16}(
"Normal" => 0,
"Low saturation" => 1,
"High saturation" => 2
),
LibExif.EXIF_TAG_CONTRAST => Dict{String,UInt16}(
"Normal" => 0,
"Soft" => 1,
"Hard" => 2
),
LibExif.EXIF_TAG_SHARPNESS => Dict{String,UInt16}(
"Normal" => 0,
"Soft" => 1,
"Hard" => 2
),
LibExif.EXIF_TAG_EXPOSURE_PROGRAM => Dict{String,UInt16}(
"Not defined" => 0,
"Manual" => 1,
"Normal program" => 2,
"Normal" => 2,
"Aperture priority" => 3,
"Aperture" => 3,
"Shutter priority" => 4,
"Shutter" => 4,
"Creative program (biased toward depth of field)" => 5,
"Creative" => 5,
"Creative program (biased toward fast shutter speed)" => 6,
"Action" => 6,
"Portrait mode (for closeup photos with the background out of focus" => 7,
"Portrait" => 7,
"Landscape mode (for landscape photos with the background in focus)" => 8,
"Landscape" => 8,
)
)
| ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | code | 7358 | const FILE_BYTE_ORDER = LibExif.EXIF_BYTE_ORDER_INTEL
"""
init_tag(exif, ifd, tag)
Initialize the entry of `tag` in `ifd` of `exif`.
"""
function init_tag(exif, ifd, tag)
exif1 = unsafe_load(exif)
entry = LibExif.exif_content_get_entry(exif1.ifd[ifd], tag)
if entry == C_NULL
entry = LibExif.exif_entry_new()
entry.tag = tag
LibExif.exif_content_add_entry(exif1.ifd[ifd], entry)
LibExif.exif_entry_initialize(entry, tag)
LibExif.exif_entry_unref(entry)
end
return entry
end
function get_ascii_buffer(ptrentry, tagv)
len = sizeof(tagv) + 1
unsafe_store!(ptrentry.size, Cuint(len), 1)
unsafe_store!(ptrentry.components, Culong(len), 1)
mem = LibExif.exif_mem_new_default()
buf = LibExif.exif_mem_alloc(mem, len)
unsafe_copyto!(buf, pointer(Vector{UInt8}(tagv * "\0")), len)
return buf
end
"""
set_value(ptrentry, tagv)
Set the value of the entry pointed by `ptrentry` to `tagv`.
"""
function set_value(ptrentry, tagv)
entry = unsafe_load(ptrentry)
if entry.format == LibExif.EXIF_FORMAT_SHORT
if entry.tag in keys(TagsDict)
LibExif.exif_set_short(entry.data,LibExif.EXIF_BYTE_ORDER_INTEL, TagsDict[entry.tag][tagv])
elseif entry.tag == LibExif.EXIF_TAG_YCBCR_SUB_SAMPLING
val = split(tagv, ",")
LibExif.exif_set_short(entry.data,LibExif.EXIF_BYTE_ORDER_INTEL, parse(Int, val[1]))
LibExif.exif_set_short(entry.data + 4,LibExif.EXIF_BYTE_ORDER_INTEL, parse(Int, val[2]))
else
@info entry.tag tagv
LibExif.exif_set_short(entry.data,LibExif.EXIF_BYTE_ORDER_INTEL, parse(Int, tagv))
end
elseif entry.format == LibExif.EXIF_FORMAT_LONG
LibExif.exif_set_long(entry.data, FILE_BYTE_ORDER, parse(Cuint, tagv))
elseif entry.format == LibExif.EXIF_FORMAT_RATIONAL
if entry.tag in (LibExif.EXIF_TAG_FNUMBER, LibExif.EXIF_TAG_APERTURE_VALUE, LibExif.EXIF_TAG_MAX_APERTURE_VALUE)
p = Rational(parse(Float32, split(tagv, "/")[2]))
else
p = rationalize(parse(Float32, tagv);tol=0.1)
end
LibExif.exif_set_rational(entry.data,FILE_BYTE_ORDER, LibExif.ExifRational(p.num, p.den))
elseif entry.format == LibExif.EXIF_FORMAT_ASCII
ptrentry.data = get_ascii_buffer(ptrentry, tagv)
elseif entry.format == LibExif.EXIF_FORMAT_SRATIONAL
p = Rational(parse(Float32, tagv))
LibExif.exif_set_srational(entry.data, FILE_BYTE_ORDER, LibExif.ExifSRational(p.num, p.den))
elseif entry.format == LibExif.EXIF_FORMAT_UNDEFINED
if entry.tag == LibExif.EXIF_TAG_FLASH_PIX_VERSION
data = Dict{String,String}(
"FlashPix Version 1.0" => "0100\0",
"FlashPix Version 1.01" => "0101\0",
"Unknown FlashPix Version" => "0000\0",
)
unsafe_copyto!(entry.data, pointer(Vector{UInt8}(data[tagv])), 5)
elseif entry.tag == LibExif.EXIF_TAG_USER_COMMENT
ptrentry.data = get_ascii_buffer(ptrentry, tagv)
else
@debug "Tag unsupported (EXIF_FORMAT_UNDEFINED)" entry.tag
end
else
@debug "Tag unsupported" entry.tag
end
end
"""
create_exif_data(tags::Dict{String, String})
Create an exif data structure from a dictionary of tags.
"""
function create_exif_data(tags)
exif = LibExif.exif_data_new()
LibExif.exif_data_set_option(exif, LibExif.EXIF_DATA_OPTION_FOLLOW_SPECIFICATION)
LibExif.exif_data_set_data_type(exif, LibExif.EXIF_DATA_TYPE_COMPRESSED)
LibExif.exif_data_set_byte_order(exif, LibExif.EXIF_BYTE_ORDER_INTEL)
LibExif.exif_data_fix(exif)
inputs = keys(tags)
for i in inputs
key = normalize_exif_flag(i)
x = LibExif.EXIF_DATA_TYPE_UNCOMPRESSED_CHUNKY
# indentify which ifds tag goes in
ifds = [
LibExif.EXIF_IFD_0,
LibExif.EXIF_IFD_1,
LibExif.EXIF_IFD_EXIF,
LibExif.EXIF_IFD_GPS,
LibExif.EXIF_IFD_INTEROPERABILITY,
LibExif.EXIF_IFD_COUNT,
]
A = [LibExif.exif_tag_get_support_level_in_ifd(key, i, x) for i in ifds]
ifd = findfirst(==(LibExif.EXIF_SUPPORT_LEVEL_MANDATORY), A)
if ifd === nothing
ifd = findfirst(==(LibExif.EXIF_SUPPORT_LEVEL_OPTIONAL), A)
end
if key == LibExif.EXIF_TAG_YCBCR_POSITIONING
ifd = 1
end
if key in (LibExif.EXIF_TAG_PIXEL_X_DIMENSION, LibExif.EXIF_TAG_PIXEL_Y_DIMENSION)
ifd = 3
end
if ifd === nothing
@debug "Tag not supported currently or No Appropriate IFD found " key
continue
end
entry = init_tag(exif, ifd, key)
set_value(entry, tags[i])
end
return exif
end
"""
write_tags(filepath::AbstractString; img::AbstractArray, tags::Dict{String,String})
Write EXIF tags to a filepath(currently support for jpeg and jpg available).
### Keyword Arguments
- `filepath::AbstractString` : Name of the file to which image and exif is written.
- `img::AbstractArray` : Image Array whose exif data is being written to the filepath mentioned above.
- `tags::Dict{String,String}` : EXIF tags and their corresponding values as defined in libexif library
### Examples
```jl
julia> using ExifViewer, TestImages
julia> img = testimage("mandrill")
julia> tags = Dict{String, String}(
"EXIF_TAG_MAKE"=>"Canon",
"EXIF_TAG_ORIENTATION"=>"Top-left",
"EXIF_TAG_X_RESOLUTION"=>"300",
"EXIF_TAG_Y_RESOLUTION"=>"300",
)
julia> write_tags("test.jpg"; img, tags)
julia> read_tags("test.jpg")
Dict{String, String} with 10 entries:
"EXIF_TAG_COLOR_SPACE" => "Uncalibrated"
"EXIF_TAG_COMPONENTS_CONFIGURATION" => "Y Cb Cr -"
"EXIF_TAG_FLASH_PIX_VERSION" => "FlashPix Version 1.0"
"EXIF_TAG_Y_RESOLUTION" => "300"
"EXIF_TAG_ORIENTATION" => "Top-left"
"EXIF_TAG_EXIF_VERSION" => "Exif Version 2.1"
"EXIF_TAG_RESOLUTION_UNIT" => "Inch"
"EXIF_TAG_MAKE" => "Canon"
"EXIF_TAG_YCBCR_POSITIONING" => "Centered"
"EXIF_TAG_X_RESOLUTION" => "300"
```
Note: some tags are present by default like EXIF version, FLASHPIX version etc as can be seen in example above.
"""
function write_tags(filepath::AbstractString; img::AbstractArray, tags::Dict{String,String})
# restricting filetype to .jpeg and .jpg
if (!(splitext(filepath)[2] in (".jpeg", ".jpg")))
throw(DomainError("Currently only jpeg and jpg files are supported for EXIF write operation."))
end
data = jpeg_encode(img)
exif = create_exif_data(tags)
exif_header = Vector{Cuchar}([0xff, 0xd8, 0xff, 0xe1])
exif_data = Ref{Ptr{Cuchar}}()
exif_data_len = Cuint(length(exif_data))
ref_exif_data_len = Ref(exif_data_len)
LibExif.exif_data_save_data(exif, exif_data, ref_exif_data_len)
groups_vec = unsafe_wrap(Array, exif_data[], 5000)
len = findfirst([0xff], groups_vec)[1]
groups_vec = groups_vec[1:max(len, 1)]
open(filepath, "w") do file
write(file, exif_header) # done
write(file, UInt8((len + 2) >> 8))
write(file, UInt8((len + 2) & 0xff))
write(file, groups_vec)
write(file, data[3:end])
end
end
| ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | code | 4860 | @testset "exifviewer.jl" begin
@testset "Basic IO" begin
open(filepath, "r") do io
@test read_tags(io; read_all=true)["EXIF_TAG_PIXEL_X_DIMENSION"] == "3000"
end
io = IOBuffer()
try
write(io, read(filepath))
@test read_tags(take!(io); read_all=true)["EXIF_TAG_PIXEL_X_DIMENSION"] == "3000"
finally
close(io)
end
@test read_tags(filepath; read_all=true)["EXIF_TAG_EXIF_VERSION"] == "Exif Version 2.1"
open(filepath, "r") do file
@test read_tags(file; read_all=true)["EXIF_TAG_ORIENTATION"] == "Top-left"
end
open(filepath, "r") do io
@test read_tags(io; tags = ["EXIF_TAG_EXIF_VERSION"])["EXIF_TAG_EXIF_VERSION"] == "Exif Version 2.1"
end
@test read_tags(filepath; tags = ["EXIF_TAG_EXIF_VERSION"])["EXIF_TAG_EXIF_VERSION"] == "Exif Version 2.1"
open(filepath, "r") do io
@test read_tags(io; tags = ["EXIF_TAG_EXIF_VERSION"])["EXIF_TAG_EXIF_VERSION"] == "Exif Version 2.1"
end
@test typeof(read_tags([0x00, 0x01])) == Dict{String, String} # to see behavior when garbage data in
end
@testset "Different IFDs" begin
@test length(read_tags(filepath; read_all=true, ifds = 1)) == 6
@test length(read_tags(filepath; read_all=true, ifds = 1:2)) == 7
@test length(read_tags(filepath; read_all=true, ifds = (1, 2))) == 7
@test length(read_tags(filepath; read_all=false, tags = ["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"])) == 2
@test length(read_tags(filepath; read_all=false, tags = ["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"], ifds = 1),) == 1
@test length(read_tags(filepath; read_all=false, tags = ["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"], ifds = 1:4),) == 2
@test length(read_tags(filepath; read_all=false, tags = ["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"], ifds = 4:5),) == 0
@test length(read_tags(filepath; read_all=false, tags = ["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"], ifds = (1, 2, 3)),) == 2
@test length(read_tags(filepath; read_all=false, tags = ["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"], ifds = (4, 5)),) == 0
@test length(read_tags(filepath; read_all=false, tags = ["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"], ifds = 1)) == 1
@test length(read_tags(filepath; read_all=false, tags = ["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"], ifds = (1, 2, 3))) == 2
# all wrongs
@test_throws BoundsError read_tags(filepath; read_all=true, ifds = 6)
@test_throws BoundsError read_tags(filepath; read_all=true, ifds = 6:7)
@test_throws BoundsError read_tags(filepath; read_all=true, ifds = (6, 7, 8))
# some right some wrongs
@test_throws BoundsError read_tags(filepath; read_all=true, ifds = -1)
@test_throws BoundsError read_tags(filepath; read_all=true, ifds = -1:6)
@test_throws BoundsError read_tags(filepath; read_all=true, ifds = (-1, 2, 3))
@test_throws BoundsError read_tags(filepath; ifds = 6)
@test_throws BoundsError read_tags(filepath; ifds = 6:7)
@test_throws BoundsError read_tags(filepath; ifds = (6, 7, 8))
@test_throws BoundsError read_tags(filepath; ifds = -1)
@test_throws BoundsError read_tags(filepath; ifds = -1:6)
@test_throws BoundsError read_tags(filepath; ifds = (-1, 2, 3))
end
@testset "Thumbnail Data" begin
ed_ptr = LE.exif_data_new_from_file(filepath)
if (ed_ptr == C_NULL)
return error("Unable to read EXIF data: invalid pointer")
end
ed = unsafe_load(ed_ptr)
tagsinfo , thumbnaildata = read_tags(filepath; tags = ["EXIF_TAG_ARTIST"], extract_thumbnail = true)
@test length(thumbnaildata) == ed.size
tagsinfo, thumbnaildata = read_tags(filepath; read_all=true, extract_thumbnail = true)
@test length(thumbnaildata) == ed.size
end
@testset "Mnote Data" begin
@test length(read_tags(get_example("canon", 1); read_mnote=true)) == 115
@test length(read_tags(get_example("fuji",1); read_mnote=true)) == 68
@test length(read_tags(get_example("olympus", 2); read_mnote=true)) == 67
@test length(read_tags(get_example("olympus", 3); read_mnote=true)) == 53
@test length(read_tags(get_example("olympus", 4); read_mnote=true)) == 48
@test length(read_tags(get_example("olympus", 5); read_mnote=true)) == 73
@test length(read_tags(get_example("pentax", 2); read_mnote=true)) == 58
@test length(read_tags(get_example("pentax", 3); read_mnote=true)) == 77
@test length(read_tags(get_example("pentax", 4); read_mnote=true)) == 63
end
end
| ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | code | 360 | @testset "libexif.jl" begin
@testset "Types check" begin
data_ptr = LE.exif_data_new_from_file(filepath)
@test typeof(data_ptr) == Ptr{LE._ExifData}
@test data_ptr != C_NULL
data = unsafe_load(data_ptr)
@test typeof(data) === LE._ExifData
@test typeof(data.ifd) == NTuple{5, Ptr{LE._ExifContent}}
end
end | ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | code | 398 | using ExifViewer
import ExifViewer.LibExif as LE
using Test, TestImages
using Downloads
filepath = testimage("earth_apollo17.jpg",download_only=true)
_wrap(name, num) = "https://github.com/ashwani-rathee/exif-sampleimages/blob/main/$(name)_makernote_variant_$num.jpg?raw=true"
get_example(x, y) = Downloads.download(_wrap(x, y))
include("libexif.jl")
include("exifviewer.jl")
include("write.jl") | ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | code | 2133 | @testset "write-exif.jl" begin
img = testimage("mandrill")
tags = Dict{String, String}(
"EXIF_TAG_MAKE"=>"Canon",
"EXIF_TAG_ARTIST"=>"Ashwani",
"EXIF_TAG_MODEL"=>"R70",
"EXIF_TAG_ORIENTATION"=>"Top-left",
"EXIF_TAG_X_RESOLUTION"=>"300",
"EXIF_TAG_Y_RESOLUTION"=>"300",
"EXIF_TAG_RESOLUTION_UNIT"=>"Centimeter",
"EXIF_TAG_FOCAL_PLANE_RESOLUTION_UNIT"=>"Inch",
"EXIF_TAG_YCBCR_POSITIONING"=>"Co-sited",
"EXIF_TAG_COMPRESSION"=>"JPEG compression",
"EXIF_TAG_FNUMBER"=>"f/2.8",
"EXIF_TAG_EXIF_VERSION"=> "Exif Version 2.1",
"EXIF_TAG_METERING_MODE"=>"Pattern",
"EXIF_TAG_FLASH"=>"Flash fired",
"EXIF_TAG_FLASH_PIX_VERSION"=> "FlashPix Version 1.0",
"EXIF_TAG_COLOR_SPACE"=>"sRGB",
"EXIF_TAG_PIXEL_Y_DIMENSION"=>"2",
"EXIF_TAG_PIXEL_X_DIMENSION"=>"2",
"EXIF_TAG_FOCAL_PLANE_X_RESOLUTION"=>"4.5",
"EXIF_TAG_FOCAL_PLANE_Y_RESOLUTION"=>"4.5",
"EXIF_TAG_SENSING_METHOD"=>"One-chip color area sensor",
"EXIF_TAG_SUBJECT_DISTANCE_RANGE"=>"Close view",
"EXIF_TAG_PLANAR_CONFIGURATION"=>"Planar format",
"EXIF_TAG_PHOTOMETRIC_INTERPRETATION"=>"CieLAB",
"EXIF_TAG_CUSTOM_RENDERED"=>"Normal process",
"EXIF_TAG_EXPOSURE_MODE"=>"Auto exposure",
"EXIF_TAG_WHITE_BALANCE"=>"Auto white balance",
"EXIF_TAG_SCENE_CAPTURE_TYPE"=>"Standard",
"EXIF_TAG_GAIN_CONTROL"=>"Normal",
"EXIF_TAG_SATURATION"=>"Normal",
"EXIF_TAG_CONTRAST"=>"Normal",
"EXIF_TAG_SHARPNESS"=>"Normal",
"EXIF_TAG_COMPONENTS_CONFIGURATION"=> "Y Cb Cr -",
"EXIF_TAG_USER_COMMENT"=>"Dummy comment",
# "EXIF_TAG_MAKER_NOTE"=>"Maker Note", # TODO: support `EXIF_TAG_MAKER_NOTE`
)
path = joinpath(tempdir(), "tmp.jpg")
write_tags(path; img, tags)
# currently only .jpg supported, different value of these were already checked
# case where key in dict is not found needs to be handled,
# support level issue needs to be handled
@test read_tags(path) == tags
rm(path)
end | ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | docs | 5228 |

<p style="text-align: center;">
ExifViewer.jl is a Julia wrapper of the C library libexif that provides EXIF support. EXIF is short for Exchangeable Image File, a format that is a standard for storing interchange information in digital photography image files using JPEG compression.
</p>
[](https://juliaimages.org/ExifViewer.jl/dev/) [](https://join.slack.com/t/julialang/shared_invite/zt-1hxxb5ryp-Ts_egJ7FRN2muQ7nkTtCNQ) [](https://opensource.org/licenses/MIT) [](https://pkgs.genieframework.com?packages=ExifViewer)
### Installation
---
If you have not yet installed Julia, please follow the [instructions](https://julialang.org/downloads/platform/) for your operating system.
Stable Version
```julia
# Enter ']' from the REPL to enter Pkg mode.
pkg> add ExifViewer
```
Dev Version
```julia
using Pkg
# Enter ']' from the REPL to enter Pkg mode.
pkg> add https://github.com/JuliaImages/ExifViewer.jl.git
```
### Usage
ExifViewer.jl provides method to read EXIF tags from images using `read_tags` methods which can
take input in form of Filepath, IO, and bytes sequence(`Vector{UInt8}`)
`read_tags` reads EXIF tags from the input source data and it returns an empty
dictionary if the source data doesn't contain EXIF tags.
There are couple of keyword arguments that are used by `read_tags` which have been
described below:
#### Keyword Arguments
- `ifds::Union{Int,NTuple,UnitRange}` : Defines which IFD(Image file directory) to search in for the EXIF tags. Default is all ifds i.e. 1:5.
- `read_all::Bool` : Defines if all EXIF tags are to be read or not. By default, `read_all` is true.
- `tags::Vector{LibExif.ExifTag}` : Defines which tags to search, in case `read_all` is false. When `read_all` is false, tags that need to be searched need to defined manually. Tags can be provided using bunch of methods but its suggested to supply a vector of strings with each string representing a EXIF tag i.e. ["`EXIF_TAG_FLASH_PIX_VERSION`", "`EXIF_TAG_ORIENTATION`"]
- `extract_thumbnail::Bool` : Defines whether to read the thumbnail data or not. By default, `extract_thumbnail` is false.
- `read_mnote::Bool` : Defines whether to read the mnote(MakerNote) tags data or not. By default, `read_mnote` is false.
List of all available tags to search is available here: https://libexif.github.io/internals/exif-tag_8h.html
#### Examples
```jl
julia> using TestImages, ExifViewer
julia> filepath = testimage("earth_apollo17.jpg", download_only=true)
julia> io = open(filepath, "r")
julia> read_tags(io; read_all=false, tags=["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"])
Dict{String, String} with 2 entries:
"EXIF_TAG_FLASH_PIX_VERSION" => "FlashPix Version 1.0"
"EXIF_TAG_ORIENTATION" => "Top-left"
julia> read_tags(filepath; read_all=false, tags=["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"])
Dict{String, String} with 2 entries:
"EXIF_TAG_FLASH_PIX_VERSION" => "FlashPix Version 1.0"
"EXIF_TAG_ORIENTATION" => "Top-left"
julia> data = read(filepath)
julia> read_tags(data, read_all=false, tags=["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"])
Dict{String, String} with 2 entries:
"EXIF_TAG_FLASH_PIX_VERSION" => "FlashPix Version 1.0"
"EXIF_TAG_ORIENTATION" => "Top-left"
```
Method to write exif data to files is also provided using `write_tags` and it writes EXIF tags to a
filepath(currently support for jpeg and jpg available).
#### Keyword Arguments
- `filepath::AbstractString` : Name of the file to which image and exif is written.
- `img::AbstractArray` : Image Array whose exif data is being written to the filepath mentioned above.
- `tags::Dict{String,String}` : EXIF tags and their corresponding values as defined in libexif library
#### Examples
```jl
julia> using ExifViewer, TestImages
julia> img = testimage("mandrill")
julia> tags = Dict{String, String}(
"EXIF_TAG_MAKE"=>"Canon",
"EXIF_TAG_ORIENTATION"=>"Top-left",
"EXIF_TAG_X_RESOLUTION"=>"300",
"EXIF_TAG_Y_RESOLUTION"=>"300",
)
julia> write_tags("test.jpg"; img, tags)
julia> read_tags("test.jpg")
Dict{String, String} with 10 entries:
"EXIF_TAG_COLOR_SPACE" => "Uncalibrated"
"EXIF_TAG_COMPONENTS_CONFIGURATION" => "Y Cb Cr -"
"EXIF_TAG_FLASH_PIX_VERSION" => "FlashPix Version 1.0"
"EXIF_TAG_Y_RESOLUTION" => "300"
"EXIF_TAG_ORIENTATION" => "Top-left"
"EXIF_TAG_EXIF_VERSION" => "Exif Version 2.1"
"EXIF_TAG_RESOLUTION_UNIT" => "Inch"
"EXIF_TAG_MAKE" => "Canon"
"EXIF_TAG_YCBCR_POSITIONING" => "Centered"
"EXIF_TAG_X_RESOLUTION" => "300"
```
Note: Some tags are present by default like EXIF version, FLASHPIX version etc as can be seen in example above.
### Contributions and Issues:
If you have questions about ExifViewer.jl, feel free to get in touch via Slack or open an issue :hearts:
| ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | docs | 4082 | ```@meta
CurrentModule = ExifViewer
```
# ExifViewer
This is the documentation for [ExifViewer](https://github.com/JuliaImages/ExifViewer.jl)
ExifViewer.jl is a Julia wrapper of the C library libexif that provides EXIF support.
# Usage
We provide method to read EXIF tags from images using `read_tags` methods which can
take input in form of Filepath, IO, and bytes sequence(`Vector{UInt8}`)
`read_tags` reads EXIF tags from the input source data and it returns an empty
dictionary if the source data doesn't contain EXIF tags.
There are couple of keyword arguments that are used by `read_tags` which have been
described below:
#### Keyword Arguments
- `ifds::Union{Int,NTuple,UnitRange}` : Defines which IFD(Image file directory) to search in for the EXIF tags. Default is all ifds i.e. 1:5.
- `read_all::Bool` : Defines if all EXIF tags are to be read or not. By default, `read_all` is true.
- `tags::Vector{LibExif.ExifTag}` : Defines which tags to search, in case `read_all` is false. When `read_all` is false, tags that need to be searched need to defined manually. Tags can be provided using bunch of methods but its suggested to supply a vector of strings with each string representing a EXIF tag i.e. ["`EXIF_TAG_FLASH_PIX_VERSION`", "`EXIF_TAG_ORIENTATION`"]
- `extract_thumbnail::Bool` : Defines whether to read the thumbnail data or not. By default, `extract_thumbnail` is false.
- `read_mnote::Bool` : Defines whether to read the mnote(MakerNote) tags data or not. By default, `read_mnote` is false.
List of all available tags to search is available here: https://libexif.github.io/internals/exif-tag_8h.html
#### Examples
```jl
julia> using TestImages, ExifViewer
julia> filepath = testimage("earth_apollo17.jpg", download_only=true)
julia> io = open(filepath, "r")
julia> read_tags(io; read_all=false, tags=["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"])
Dict{Any, Any} with 2 entries:
"EXIF_TAG_FLASH_PIX_VERSION" => "FlashPix Version 1.0"
"EXIF_TAG_ORIENTATION" => "Top-left"
julia> read_tags(filepath; read_all=false, tags=["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"])
Dict{Any, Any} with 2 entries:
"EXIF_TAG_FLASH_PIX_VERSION" => "FlashPix Version 1.0"
"EXIF_TAG_ORIENTATION" => "Top-left"
julia> data = read(filepath)
julia> read_tags(data, read_all=false, tags=["EXIF_TAG_FLASH_PIX_VERSION", "EXIF_TAG_ORIENTATION"])
Dict{Any, Any} with 2 entries:
"EXIF_TAG_FLASH_PIX_VERSION" => "FlashPix Version 1.0"
"EXIF_TAG_ORIENTATION" => "Top-left"
```
Method to write exif data to files is also provided using `write_tags` and it writes EXIF tags to a
filepath(currently support for jpeg and jpg available).
### Keyword Arguments
- `filepath::AbstractString` : Name of the file to which image and exif is written.
- `img::AbstractArray` : Image Array whose exif data is being written to the filepath mentioned above.
- `tags::Dict{String,Any}` : EXIF tags and their corresponding values as defined in libexif library
### Examples
```jl
julia> using ExifViewer, TestImages
julia> img = testimage("mandrill")
julia> tags = Dict{String, Any}(
"EXIF_TAG_MAKE"=>"Canon",
"EXIF_TAG_ORIENTATION"=>"Top-left",
"EXIF_TAG_X_RESOLUTION"=>"300",
"EXIF_TAG_Y_RESOLUTION"=>"300",
)
julia> write_tags("test.jpg"; img, tags=tags)
julia> read_tags("test.jpg")
Dict{String, Any} with 10 entries:
"EXIF_TAG_COLOR_SPACE" => "Uncalibrated"
"EXIF_TAG_COMPONENTS_CONFIGURATION" => "Y Cb Cr -"
"EXIF_TAG_FLASH_PIX_VERSION" => "FlashPix Version 1.0"
"EXIF_TAG_Y_RESOLUTION" => "300"
"EXIF_TAG_ORIENTATION" => "Top-left"
"EXIF_TAG_EXIF_VERSION" => "Exif Version 2.1"
"EXIF_TAG_RESOLUTION_UNIT" => "Inch"
"EXIF_TAG_MAKE" => "Canon"
"EXIF_TAG_YCBCR_POSITIONING" => "Centered"
"EXIF_TAG_X_RESOLUTION" => "300"
```
Note: some tags are present by default like EXIF version, FLASHPIX version etc as can be seen in example above.
```@autodocs
Modules = [ExifViewer]]
``` | ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.1.4 | cacf335f8ad079f84faf2aa3f66b61fa2971b0b8 | docs | 195 | The files in `lib/` folder automatically generated by Clang.jl, to regenerate the files:
```console
julia --project=gen -e 'using Pkg; Pkg.instantiate()'
julia --project=gen gen/generator.jl
``` | ExifViewer | https://github.com/JuliaImages/ExifViewer.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 5124 | """
This module groups all callback functions
that are called at various points during the solution process.
See their respective docstrings for more information.
Every callback takes an observer as its first argument,
which is passed via an optional keyword argument to `solve`.
During `solve(::GALEProblem, ::LyapunovSolver; observer)`:
* [`observe_gale_start!`](@ref)
* [`observe_gale_step!`](@ref)
* [`observe_gale_done!`](@ref)
* [`observe_gale_failed!`](@ref)
* [`observe_gale_metadata!`](@ref)
During `solve(::GAREProblem, ::AlgebraicRiccatiSolver; observer)`:
* [`observe_gare_start!`](@ref)
* [`observe_gare_step!`](@ref)
* [`observe_gare_done!`](@ref)
* [`observe_gare_failed!`](@ref)
During `solve(::GDREProblem, ::Algorithm; observer)`:
* [`observe_gdre_start!`](@ref)
* [`observe_gdre_step!`](@ref)
* [`observe_gdre_done!`](@ref)
# Extended help
Hook into above callbacks by first defining a custom observer type.
```julia
mutable struct ResidualObserver
norms::Vector{Float64}
abstol::Float64
ResidualObserver() = new(Float64[], -1.0)
end
```
Then, create custom methods to above callbacks.
If the observer needs to store any information,
use some global variables (not recommended),
have the observer be mutable.
Note that `Callbacks` has to be imported manually;
this is a deliberate choice.
```julia
import DifferentialRiccatiEquations.Callbacks
function Callbacks.observe_gale_step!(o::ResidualObserver, _prob, _alg, abstol::Float64, _reltol)
o.abstol = abstol
end
function Callbacks.observe_gale_step!(o::ResidualObserver, _iter, _sol, _residual, residual_norm::Float64)
push!(o.norms, residual_norm)
end
```
The observer is passed into the solution procedure as follows.
```julia
prob = GALEProblem(E, A, C)
alg = ADI()
obs = ResidualObserver()
solve(prob, alg; observer=obs)
@show obs.norms[end] <= obs.abstol
```
!!! todo
Update extended help to use doctests.
"""
module Callbacks
export observe_gale_start!,
observe_gale_step!,
observe_gale_done!,
observe_gale_failed!,
observe_gale_metadata!
export observe_gare_start!,
observe_gare_step!,
observe_gare_done!,
observe_gare_failed!,
observe_gare_metadata!
export observe_gdre_start!,
observe_gdre_step!,
observe_gdre_done!
"""
observe_gale_start!(observer, prob::GALEProblem, alg::LyapunovSolver)
Notify `observer` at the start of solving the GALE.
"""
observe_gale_start!(::Any, args...) = nothing
const COMMON_GALE_DESC = """
The observer may compute ans store any metrics of the subsequent arguments,
but it must not modify any of them.
* `X`: solution candidate
* `residual`: residual corresponding to `X`;
usually of the same data type as `X`
* `residual_norm`: internal approximation of the norm of `residual`
"""
"""
observe_gale_step!(observer, iter::Int, X, residual, residual_norm)
Notify `observer` for an iterative GALE algorithm,
that iteration number `iter` has been completed.
$COMMON_GALE_DESC
!!! note
The iterations `iter` may not be consequtive.
If an algorithm computes multiple steps at once
and has no (cheap) representation of the intermediate solution candidates,
the difference between the values of `iter`
of subsequent calls to `observe_gale_step!`
may differ by more than one.
"""
observe_gale_step!(::Any, args...) = nothing
"""
observe_gale_done!(observer, iters::Int, X, residual, residual_norm)
Notify `observer` at the end of solving the GALE.
$COMMON_GALE_DESC
"""
observe_gale_done!(::Any, args...) = nothing
"""
observe_gale_failed!(observer)
Notify `observer` that the algorithm has failed to solve the GALE.
[`observe_gale_done!`](@ref) will be called regardless.
"""
observe_gale_failed!(::Any) = nothing
"""
observe_gale_metadata!(observer, desc::String, metadata)
Notify `observer` on some `metadata` the algorithm has computed.
`desc` gives a brief description of the metadata.
### Example
The [`ADI`](@ref) calls `observe_gale_metadata!(observer, "ADI shifts", μ)`,
where `μ` are the (newly) computed ADI shift parameters.
"""
observe_gale_metadata!(::Any, args...) = nothing
"""
observe_gdre_start!(observer, ::GDREProblem, ::Algorithm)
Notify `observer` at the start of solving the GDRE.
"""
observe_gdre_start!(::Any, args...) = nothing
"""
observe_gdre_step!(observer, t::Float64, X, K)
Notify `observer` that the step to time point `t` has been completed.
* `X`: solution at time `t`
* `K`: feedback matrix `K = B' * X * E`
where `B` denotes the input map of the associated [`GDREProblem`](@ref)
"""
observe_gdre_step!(::Any, args...) = nothing
"""
observe_gdre_done!(observer)
Notify `observer` at the end of solving the GDRE.
"""
observe_gdre_done!(::Any) = nothing
# TODO: refactor callbacks to receive problem instance:
# observe_start(::Handler, ::Problem, args...)
observe_gare_start!(::Any, args...) = nothing
observe_gare_step!(::Any, args...) = nothing
observe_gare_done!(::Any, args...) = nothing
observe_gare_failed!(::Any) = nothing
observe_gare_metadata!(::Any, args...) = nothing
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 1731 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
module DifferentialRiccatiEquations
using CommonSolve: CommonSolve, solve
using Compat: @something
using LinearAlgebra
using MatrixEquations: lyapc, lyapcs!, utqu!
using UnPack: @unpack
using SparseArrays: SparseArrays,
SparseMatrixCSC,
AbstractSparseMatrixCSC,
issparse,
spzeros,
spdiagm
using TimerOutputs: @timeit_debug
include("Stuff.jl")
include("Shifts.jl")
include("Callbacks.jl")
using .Callbacks
include("LDLt.jl")
include("LowRankUpdate.jl")
include("lyapunov/types.jl")
include("lyapunov/adi.jl")
include("util/_zeros.jl")
include("util/_diagm.jl")
include("util/_dcat.jl")
include("util/_hcat.jl")
include("util/restrict.jl")
abstract type Algorithm end
struct Ros1 <: Algorithm end
struct Ros2 <: Algorithm end
struct Ros3 <: Algorithm end
struct Ros4 <: Algorithm end
include("riccati/types.jl")
include("riccati/residual.jl")
include("riccati/dense_ros1.jl")
include("riccati/dense_ros2.jl")
include("riccati/dense_ros3.jl")
include("riccati/dense_ros4.jl")
include("riccati/lowrank_ros1.jl")
include("riccati/lowrank_ros2.jl")
include("riccati/newton-adi.jl")
function CommonSolve.solve(
p::GDREProblem,
a::Algorithm;
dt::Real,
save_state::Bool=false,
observer=nothing,
kwargs...,
)
_solve(
p,
a;
dt=dt,
save_state=save_state,
observer=observer,
kwargs...,
)
end
export solve
export GDREProblem, Ros1, Ros2, Ros3, Ros4
export GAREProblem, NewtonADI
export GALEProblem, ADI
export LDLᵀ, concatenate!, compress!
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 5949 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
using Compat: allequal
"""
LDLᵀ{TL,TD}(L::TL, D::TD)
LDLᵀ{TL,TD}(Ls::Vector{TL}, Ds::Vector{TD})
A lazy representation of `L * D * L'` that supports the following functions:
* `+(::LDLᵀ, ::LDLᵀ)` and `+(::LDLᵀ{TL,TD}, ::Tuple{TL,TD})`
* `*(::Real, ::LDLᵀ)`
* `size`
* `rank` which yields the length of the inner dimension, i.e. `size(D, 1)`
* `zero` which yields a rank 0 representation
* [`concatenate!`](@ref) (expert use only)
* [`compress!`](@ref) (expert use only)
Iterating the structure yields `L::TL` and `D::TD`.
This calls [`compress!`](@ref), if necessary.
For convenience, the structure might be converted to a matrix via `Matrix`.
It is recommended to use this only for testing.
"""
struct LDLᵀ{TL,TD}
Ls::Vector{TL}
Ds::Vector{TD}
LDLᵀ(L::TL, D::TD) where {TL, TD} = new{TL,TD}([L], [D])
LDLᵀ{TL,TD}(L::TL, D::TD) where {TL, TD} = new{TL,TD}([L], [D])
LDLᵀ{TL,TD}(L::Vector{TL}, D::Vector{TD}) where {TL, TD} = new{TL,TD}(L, D)
end
Base.eltype(::Type{LDLᵀ{TL,TD}}) where {TL,TD} = promote_type(eltype(TL), eltype(TD))
# Mainly for testing
function Base.Matrix(X::LDLᵀ)
@unpack Ls, Ds = X
L = first(Ls)
n = size(L, 1)
M = zeros(eltype(L), n, n)
for (L, D) in zip(Ls, Ds)
M .+= L * D * L'
end
return M
end
# Destructuring via iteration
function Base.iterate(X::LDLᵀ)
length(X.Ls) > 1 && compress!(X)
only(X.Ls), Val(:D)
end
Base.iterate(LD::LDLᵀ, ::Val{:D}) = only(LD.Ds), nothing
Base.iterate(::LDLᵀ, _) = nothing
Base.size(X::LDLᵀ, i) = i <= 2 ? size(first(X.Ls), 1) : 1
Base.size(X::LDLᵀ) = (n = size(X, 1); (n, n))
"""
norm(::LDLᵀ)
Compute the Frobenius norm of a LDLᵀ factorization.
The technique is similar to the one described in
> Benner, Li, Penzl. Numerical solution of large-scale Lyapunov equations,
> Riccati equations, and linear-quadratic optimal control problems.
> Numerical Linear Algebra with Applications 2008. DOI: 10.1002/nla.622
"""
@timeit_debug "norm(::LDLᵀ)" function LinearAlgebra.norm(X::LDLᵀ)
# Decompose while not triggering compression.
concatenate!(X)
L = only(X.Ls)
D = only(X.Ds)
# TODO: use specialized TSQR ("tall and skinny QR") algorithm.
# TODO: evaluate whether `compress!` could share any code with `norm`.
if VERSION < v"1.7"
_, R = qr(L, Val(false)) # no pivoting
else
_, R = qr(L, NoPivot())
end
# The Q operator of the QR decomposition does not alter the Frobenius norm.
# It may therefore be omitted from the matrix inside the norm.
norm(R * D * R')
end
LinearAlgebra.rank(X::LDLᵀ) = sum(D -> size(D, 1), X.Ds)
function Base.zero(X::LDLᵀ{TL,TD}) where {TL,TD}
n = size(X, 1)
L = _zeros(TL, n, 0)
D = _zeros(TD, 0, 0)
LDLᵀ{TL,TD}(L, D)
end
function Base.:(+)(X1::LDLᵀ{TL,TD}, X2::LDLᵀ) where {TL,TD}
if (n1 = size(X1, 1)) != (n2 = size(X2, 1))
throw(DimensionMismatch("outer dimensions must match, got $n1 and $n2 instead"))
end
Ls = copy(X1.Ls)
Ds = copy(X1.Ds)
append!(Ls, X2.Ls)
append!(Ds, X2.Ds)
X = LDLᵀ{TL,TD}(Ls, Ds)
maybe_compress!(X)
end
Base.:(-)(X::LDLᵀ{TL,TD}) where {TL,TD} = LDLᵀ{TL,TD}(X.Ls, -X.Ds)
Base.:(-)(X::LDLᵀ, Y::LDLᵀ) = X + (-Y)
# TODO: Make this more efficient by storing the scalar as a field of LDLᵀ.
function Base.:(*)(α::Real, X::LDLᵀ)
L, D = X
LDLᵀ(L, α*D)
end
function compression_due(X::LDLᵀ)
# If there is only one component, it has likely already been compressed:
length(X.Ls) == 1 && return false
# Compression is due every couple of modifications:
# TODO: make this configurable
length(X.Ls) >= 10 && return true
# Compression is due if rank is too large:
# TODO: make this configurable
n = size(X, 1)
r = rank(X)
return r >= 0.5n
end
function maybe_compress!(X::LDLᵀ)
compression_due(X) || return X
compress!(X)
end
"""
concatenate!(X::LDLᵀ)
Concatenate the internal components such that `L` and `D` may be obtained via `L, D = X`.
This function is roughly equivalent to `L = foldl(hcat, X.Ls)` and `D = foldl(dcat, Ds)`,
where `dcat` is pseudo-code for "diagonal concatenation".
This is a somewhat cheap operation.
See also: [`compress!`](@ref)
"""
@timeit_debug "concatenate!(::LDLᵀ)" function concatenate!(X::LDLᵀ{TL,TD}) where {TL,TD}
@unpack Ls, Ds = X
@assert length(Ls) == length(Ds)
length(Ls) == 1 && return X
L = _hcat(TL, Ls)
D = _dcat(TD, Ds)
resize!(X.Ls, 1)
resize!(X.Ds, 1)
X.Ls[1] = L
X.Ds[1] = D
return X
end
"""
compress!(X::LDLᵀ)
Concatenate the internal components and perform a column compression following [^Lang2015].
This is an expensive operation.
See also: [`concatenate!`](@ref)
[^Lang2015]: N Lang, H Mena, and J Saak, "On the benefits of the LDLT factorization for large-scale differential matrix equation solvers" Linear Algebra and its Applications 480 (2015): 44-71. [doi:10.1016/j.laa.2015.04.006](https://doi.org/10.1016/j.laa.2015.04.006)
"""
@timeit_debug "compress!(::LDLᵀ)" function compress!(X::LDLᵀ{TL,TD}) where {TL,TD}
concatenate!(X)
L = only(X.Ls)
D = only(X.Ds)
@timeit_debug "QR" if VERSION < v"1.7"
Q, R, p = qr(L, Val(true)) # pivoting
else
Q, R, p = qr(L, ColumnNorm())
end
ip = invperm(p)
RΠᵀ = R[:,ip]
S = Symmetric(RΠᵀ*D*(RΠᵀ)')
λ, V = @timeit_debug "Eigen" eigen(S; sortby = x -> -abs(x))
# only use "large" eigenvalues,
# cf. [Kürschner2016, p. 94]
# (modified to retain negative ones)
ε = max(1, abs(λ[1])) * length(λ) * eps()
r = something(findlast(l -> abs(l) >= ε, λ), 0)
@debug "compress!(::LDLᵀ)" λ[1] λ[end] count(>(ε), λ) count(<(-ε), λ) oldrank=size(D,1) newrank=r
Vᵣ = @view V[:, 1:r]
X.Ls[1] = (Q * Vᵣ)::TL
X.Ds[1] = _diagm(TD, λ[1:r])::TD
return X
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 3266 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
"""
LowRankUpdate{TA,T,TU,TV}(A::TA, α::T, U::TU, V::TV)
Lazy representation of `A + inv(α)*U*V` that supports the following functions:
* `\\` via the Sherman-Morrison-Woodbury formula
* `+(::LowRankUpdate, ::AbstractMatrix)` to update `A`
* `adjoint` which returns a `LowRankUpdate`
* `size`
Iterating the structure produces the components `A`, `α`, `U` and `V`.
It is recommended to use [`lr_update`](@ref) to create a suitable
representation of `A + inv(α)*U*V`.
"""
struct LowRankUpdate{TA,T,TU,TV}
A::TA
α::T
U::TU
V::TV
LowRankUpdate{T,TA,TU,TV}(A, α, U, V) where {TA,T,TU,TV} = new{TA,T,TU,TV}(A, α, U, V)
LowRankUpdate(A::TA, α::T, U::TU, V::TV) where {TA,T,TU,TV} = new{TA,T,TU,TV}(A, α, U, V)
end
"""
lr_update(A::Matrix, α, U, V)
lr_update(A::AbstractSparseMatrixCSC, α, U, V)
Return a suitable representation of `A + inv(α)*U*V`.
For dense `A`, compute `A + inv(α)*U*V` directly.
For sparse `A`, return a [`LowRankUpdate`](@ref).
"""
lr_update
lr_update(A::Matrix{T}, α, U, V) where {T} = A + (inv(α)*U)*V
lr_update(A::AbstractSparseMatrixCSC, α, U, V) = LowRankUpdate(A, α, U, V)
Base.eltype(::Type{LowRankUpdate{TA,T,TU,TV}}) where {TA,T,TU,TV} = Base.promote_eltype(TA, T, TU, TV)
Base.iterate(AUV::LowRankUpdate) = AUV.A, Val(:a)
Base.iterate(AUV::LowRankUpdate, ::Val{:a}) = AUV.α, Val(:U)
Base.iterate(AUV::LowRankUpdate, ::Val{:U}) = AUV.U, Val(:V)
Base.iterate(AUV::LowRankUpdate, ::Val{:V}) = AUV.V, nothing
Base.size(AUV::LowRankUpdate) = size(AUV.A)
Base.size(AUV::LowRankUpdate, i) = size(AUV.A, i)
function Base.adjoint(AUV::LowRankUpdate)
A, α, U, V = AUV
LowRankUpdate(A', α', V', U')
end
function Matrix(AUV::LowRankUpdate)
A, α, U, V = AUV
A + inv(α) * (U * V)
end
_factorize(X) = factorize(X)
function _factorize(X::AbstractSparseMatrixCSC)
F = factorize(X)
F isa Diagonal || return F
# If `F` is a `Diagonal`, its diagonal `F.diag` will be a `SparseVector`.
# Given that `F` defines an invertible operator, its diagonal is effectively
# a dense vector. In this setting, the sparse vector type incurs a certain
# runtime overhead when solving linear systems in `F` as compared to a dense
# vector type. Get rid of this overhead:
D = Diagonal(Vector(F.diag))
return D
end
@timeit_debug "Sherman-Morrison-Woodbury" function Base.:(\)(AUV::LowRankUpdate, B::AbstractVecOrMat)
A, α, U, V = AUV
FA = @timeit_debug "factorize (sparse)" _factorize(A)
A⁻¹B = @timeit_debug "solve (sparse 1)" FA \ B
A⁻¹U = @timeit_debug "solve (sparse 2)" FA \ U
@timeit_debug "solve (dense)" begin
S = α*I + V * A⁻¹U
S⁻¹VA⁻¹B = S \ (V * A⁻¹B)
end
X = A⁻¹B - A⁻¹U*S⁻¹VA⁻¹B
return X
end
function Base.:(+)(AUV::LowRankUpdate, E::AbstractMatrix)
@assert issparse(E)
A, α, U, V = AUV
LowRankUpdate(A+E, α, U, V)
end
function Base.:(*)(AUV::LowRankUpdate, X::AbstractVecOrMat)
size(X, 1) == size(X, 2) && @warn(
"Multiplying LowRankUpdate by square matrix; memory usage may increase severely",
dim = size(X, 1),
)
A, α, U, V = AUV
A*X + inv(α)*(U*(V*X))
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 2716 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
"""
This module groups all pre-defined shift strategies.
* [`Shifts.Heuristic`](@ref)
* [`Shifts.Projection`](@ref)
* [`Shifts.Cyclic`](@ref)
* [`Shifts.Wrapped`](@ref)
# Extended help
To define a custom shift strategy,
create a mutable subtype of `Shifts.Strategy`,
define a method for [`Shifts.init`](@ref) and, optionally,
methods for [`Shifts.update!`](@ref) and [`Shifts.take!`](@ref).
```julia
struct FortyTwo <: Shifts.Strategy end
Shifts.init(::FortyTwo, _...) = FortyTwo()
Shifts.take!(::FortyTwo) = 42
```
If it is customary to generate multiple shift parameters at once,
that are then to be used one-by-one, define a method for
[`Shifts.take_many!`](@ref) and have [`Shifts.init`](@ref) return a
[`Shifts.BufferedIterator`](@ref).
```julia
struct FibonacciShifts <: Shifts.Strategy
"Number of shifts to generate at a time"
n::Int
function FibonacciShifts(n::Int)
n >= 2 || error("batch size is too small")
new(n)
end
end
struct FibonacciShiftsIterator
n::Int
f1::Int
f2::Int
end
function Shifts.init(f::FibonacciShifts)
Shifts.BufferedIterator(FibonacciShiftsIterator(f.n, 0, 1))
end
function Shifts.take_many!(it::FibonacciShiftsIterator)
n = it.n
# Generate n shifts at once:
f = Vector{Int}(undef, n)
f[1] = it.f1
f[2] = it.f2
for i in 3:n
f[i] = f[i-1] + f[i-2]
end
# Prepare next batch:
it.f1 = f[end-1] + f[end]
it.f2 = f[end] + it.f1
return f
end
```
"""
module Shifts
export Cyclic, Wrapped
export Heuristic, Projection
abstract type Strategy end
"""
Shifts.init(::Shifts.Strategy, prob)
Create and initialize a shift generator from problem data.
The returned iterator will immediately be [`Shifts.update!`](@ref)ed
with initial guess and residual of the iteration.
"""
init
"""
Shifts.update!(shifts, X, R, Vs...)
Pass most recent solution update to shift generator `shifts`.
* `X`: current solution candidate
* `R`: outer factor of residual corresponding to `X`
* `Vs`: outer factors of most recent updates comprising `X`
This operation must be cheap.
Defer the computation of new shift parameters to [`Shifts.take!`](@ref) or [`Shifts.take_many!`](@ref).
Default: no-op.
"""
update!(_, _, _, _...) = nothing
"""
Shifts.take!(shifts)
Return the next shift parameter from shift generator `shifts`.
This operation may be expensive.
Compute new shift parameters, if needed.
Default: `popfirst!(shifts)`
"""
take!(shifts) = popfirst!(shifts)
using ..Stuff
include("shifts/helpers.jl")
include("shifts/heuristic.jl")
include("shifts/projection.jl")
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 967 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
module Stuff
using SparseArrays, LinearAlgebra
export orth, restrict
restrict(A::AbstractMatrix, Q) = Q' * A * Q
orth(N::SparseMatrixCSC) = orth(Matrix(N))
function orth(N::Matrix{T}) where {T}
if VERSION < v"1.7"
QR = qr(N, Val(true)) # pivoted
else
QR = qr(N, ColumnNorm())
end
R = QR.R
# TODO: Find reference! As of LAPACK 3.1.2 or so,
# the diagonal of R is sorted with decreasing absolute value,
# and R is diagonal dominant. Therefore, it may be used to discover the rank.
# Note that column permutations don't matter for span(N) == span(Q).
ε = size(N, 1) * eps()
r = 0
for outer r in 1:size(R, 1)
abs(R[r,r]) > ε && continue
r -= 1
break
end
Q = zeros(T, size(N, 1), r)
for i in 1:r
Q[i,i] = 1
end
lmul!(QR.Q, Q)
return Q
end
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 3689 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
using Compat: @something
function CommonSolve.solve(
prob::GALEProblem{LDLᵀ{TL,TD}},
::ADI;
initial_guess::Union{Nothing,LDLᵀ{TL,TD}}=nothing,
maxiters=100,
reltol=size(prob.A, 1) * eps(),
abstol=reltol * norm(prob.C), # use same tolerance as if initial_guess=zero(C)
observer=nothing,
shifts::Shifts.Strategy=Shifts.Projection(2),
) where {TL,TD}
@timeit_debug "callbacks" observe_gale_start!(observer, prob, ADI())
initial_guess = @something initial_guess zero(prob.C)
@unpack E, A, C = prob
# Compute initial residual
X::LDLᵀ{TL,TD} = initial_guess::LDLᵀ{TL,TD}
R::TL, T::TD = initial_residual = residual(prob, X)::LDLᵀ{TL,TD}
initial_residual_norm = norm(initial_residual)
# Initialize shifts
@timeit_debug "shifts" begin
shifts = Shifts.init(shifts, prob)
Shifts.update!(shifts, X, R)
end
# Perform actual ADI
i = 1
local V, V₁, V₂ # ADI increments
local ρR # norm of residual
@timeit_debug "callbacks" observe_gale_step!(observer, 0, X, initial_residual, initial_residual_norm)
while true
μ = @timeit_debug "shifts" Shifts.take!(shifts)
@timeit_debug "callbacks" observe_gale_metadata!(observer, "ADI shifts", μ)
# Continue with ADI:
Y = (-2real(μ) * T)::TD
if isreal(μ)
μᵢ = real(μ)
F = A' + μᵢ*E
@timeit_debug "solve (real)" V = (F \ R)::TL
X += LDLᵀ(V, Y)
R -= (2μᵢ * (E'*V))::TL
i += 1
@timeit_debug "shifts" Shifts.update!(shifts, X, R, V)
else
μ_next = @timeit_debug "shifts" Shifts.take!(shifts)
@assert μ_next ≈ conj(μ)
@timeit_debug "callbacks" observe_gale_metadata!(observer, "ADI shifts", μ_next)
μᵢ = μ
F = A' + μᵢ*E
@timeit_debug "solve (complex)" V = F \ R
δ = real(μᵢ) / imag(μᵢ)
Vᵣ = real(V)
Vᵢ = imag(V)
V′ = Vᵣ + δ*Vᵢ
V₁ = √2 * V′
V₂ = sqrt(2δ^2 + 2) * Vᵢ
X = X + LDLᵀ(V₁, Y) + LDLᵀ(V₂, Y)
R -= (4real(μ) * (E'*V′))::TL
i += 2
@timeit_debug "shifts" Shifts.update!(shifts, X, R, V₁, V₂)
end
residual = LDLᵀ(R, T)
ρR = norm(residual)
@timeit_debug "callbacks" observe_gale_step!(observer, i-1, X, residual, ρR)
@debug "ADI" i rank(X) residual=ρR
ρR <= abstol && break
if i > maxiters
@timeit_debug "callbacks" observe_gale_failed!(observer)
@warn "ADI did not converge" residual=ρR abstol maxiters
break
end
end
_, D = X # run compression, if necessary
iters = i - 1 # actual number of ADI steps performed
@debug "ADI done" i=iters maxiters residual=ρR abstol rank(X) rank_initial_guess=rank(initial_guess) rank_rhs=rank(C) rank_residual=size(R)
@timeit_debug "callbacks" observe_gale_done!(observer, iters, X, LDLᵀ(R, T), ρR)
return X
end
@timeit_debug "residual(::GALEProblem, ::LDLᵀ)" function residual(
prob::GALEProblem{LDLᵀ{TL,TD}},
val::LDLᵀ{TL,TD},
) where {TL,TD}
@unpack E, A, C = prob
G, S = C
L, D = val
n_G = size(G, 2)
n_0 = size(L, 2)
dim = n_G + 2n_0
dim == n_G && return C
R::TL = _hcat(TL, G, E'L, A'L)
T::TD = _zeros(TD, dim, dim)
i1 = 1:n_G
i2 = (1:n_0) .+ n_G
i3 = i2 .+ n_0
T[i1, i1] = S
T[i3, i2] = D
T[i2, i3] = D
R̃ = LDLᵀ(R, T)::LDLᵀ{TL,TD}
compress!(R̃) # unconditionally
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 385 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
"""
Generalized algebraic Lyapunov equation
A'XE + E'XA = -C
having the fields `A`, `E` and `C`.
"""
struct GALEProblem{T}
E
A
C::T
GALEProblem(E, A, C::T) where {T} = new{T}(E, A, C)
end
abstract type LyapunovSolver end
struct ADI <: LyapunovSolver end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 1133 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
function _solve(
prob::GDREProblem{<:Matrix},
alg::Ros1;
dt::Real,
save_state::Bool,
observer,
)
observe_gdre_start!(observer, prob, alg)
@unpack E, A, B, C, tspan = prob
Ed = collect(E)
X = prob.X0
tstops = tspan[1]:dt:tspan[2]
len = length(tstops)
# Output Trajectories
Xs = [X]
save_state && sizehint!(Xs, len)
K = (B'*X)*E
Ks = [K]
sizehint!(Ks, len)
observe_gdre_step!(observer, tstops[1], X, K)
for i in 2:len
τ = tstops[i-1] - tstops[i]
# Coefficient Matrix of the Lyapunov Equation
F = (A-B*K) - E/(2τ)
R = C'*C + K'*K + (1/τ)*E'*X*E
# Only for safety
R = real(R+R')/2
# Update X
X = lyapc(F', Ed', R)
save_state && push!(Xs, X)
# Update K
K = (B'*X)*E
push!(Ks, K)
observe_gdre_step!(observer, tstops[i], X, K)
end
save_state || push!(Xs, X)
observe_gdre_done!(observer)
return DRESolution(Xs, Ks, tstops)
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 1681 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
function _solve(
prob::GDREProblem{<:Matrix},
alg::Ros2;
dt::Real,
save_state::Bool,
observer,
)
observe_gdre_start!(observer, prob, alg)
@unpack E, A, B, C, tspan = prob
Ed = collect(E)
X = prob.X0
tstops = tspan[1]:dt:tspan[2]
len = length(tstops)
# Global parameter for the method
γ = 1+(1/sqrt(2))
# Output Trajectories
Xs = [X]
save_state && sizehint!(Xs, len)
K = (B'*X)*E
Ks = [K]
sizehint!(Ks, len)
observe_gdre_step!(observer, tstops[1], X, K)
CᵀC = C'C
for i in 2:len
τ = tstops[i-1] - tstops[i]
gF = γ*τ*(A-B*K) - E/2
Fs, Es, Q, Z = schur(gF, Ed)
# Solve Lyapunov equation of 1st stage
AᵀXE = (A' * X) * E
R = CᵀC + AᵀXE + (AᵀXE)' - K'K
R = real(R+R')/2
utqu!(R, Z) # R = Z'*R*Z
lyapcs!(Fs, Es, R; adj=true)
K1 = R
utqu!(K1, Q') # K1 = Q*K1*Q'
# Solve Lyapunov equation of 2nd stage
BᵀK₁E = (B' * K1) * E
R2 = (-τ^2 * BᵀK₁E)' * BᵀK₁E - (2-1/γ)*E'*K1*E
R2 = real(R2+R2')/2
utqu!(R2, Z) # R2 = Z'*R2*Z
lyapcs!(Fs, Es, R2; adj=true)
K̃2 = R2
utqu!(K̃2, Q') # K̃2 = Q*K̃2*Q'
K2 = K̃2 + (4-1/γ)*K1
# Update X
X = X + (τ/2)*K2
save_state && push!(Xs, X)
# Update K
K = (B'*X)*E
push!(Ks, K)
observe_gdre_step!(observer, tstops[i], X, K)
end
save_state || push!(Xs, X)
observe_gdre_done!(observer)
return DRESolution(Xs, Ks, tstops)
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 2136 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
function _solve(
prob::GDREProblem{<:Matrix},
alg::Ros3;
dt::Real,
save_state::Bool,
observer,
)
observe_gdre_start!(observer, prob, alg)
@unpack E, A, B, C, tspan = prob
Ed = collect(E)
X = prob.X0
tstops = tspan[1]:dt:tspan[2]
len = length(tstops)
# Output Trajectories
Xs = [X]
save_state && sizehint!(Xs, len)
K = (B'*X)*E
Ks = [K]
sizehint!(Ks, len)
observe_gdre_step!(observer, tstops[1], X, K)
# Global parameter for the method
γ = 7.886751345948129e-1
a21 = 1.267949192431123
c21 = -1.607695154586736
c31 = -3.464101615137755
c32 = -1.732050807568877
m1 = 2
m2 = 5.773502691896258e-1
m3 = 4.226497308103742e-1
CᵀC = C'C
for i in 2:len
τ = tstops[i-1] - tstops[i]
gF = (A - B*K) - E/(2γ*τ)
Fs, Es, Q, Z = schur(gF, Ed)
# Solve Lyapunov equation of 1st stage
AXE = A'X*E
R = CᵀC + AXE + AXE' - K'K
R = real(R+R')/2
utqu!(R, Z) # R = Z'*R*Z
lyapcs!(Fs, Es, R; adj=true)
K1 = R
utqu!(K1, Q') # K1 = Q*K1*Q'
# Solve Lyapunov equation of 2nd stage
RX = (A'K1 - K'*(B'K1))*E
R23 = a21*(RX+RX')
R2 = R23 + (c21/τ)*E'K1*E
R2 = real(R2+R2')/2
utqu!(R2, Z) # R2 = Z'*R2*Z
lyapcs!(Fs, Es, R2; adj=true)
K21 = R2
utqu!(K21, Q') # K21 = Q*K21*Q'
# Solve Lyapunov equation of 3rd stage
R3 = R23 + E'*(((c31/τ)+(c32/τ))*K1 + (c32/τ)*K21)*E
R3 = real(R3+R3')/2
utqu!(R3, Z) # R3 = Z'*R3*Z
lyapcs!(Fs, Es, R3; adj=true)
K31 = R3
utqu!(K31, Q') # K31 = Q*K31*Q'
# Update X
X = X + (m1+m2+m3)*K1 + m2*K21 + m3*K31
save_state && push!(Xs, X)
# Update K
K = (B'*X)*E
push!(Ks, K)
observe_gdre_step!(observer, tstops[i], X, K)
end
save_state || push!(Xs, X)
observe_gdre_done!(observer)
return DRESolution(Xs, Ks, tstops)
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 2430 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
function _solve(
prob::GDREProblem{<:Matrix},
alg::Ros4;
dt::Real,
save_state::Bool,
observer,
)
observe_gdre_start!(observer, prob, alg)
@unpack E, A, B, C, tspan = prob
Ed = collect(E)
X = prob.X0
tstops = tspan[1]:dt:tspan[2]
len = length(tstops)
# Output Trajectories
Xs = [X]
save_state && sizehint!(Xs, len)
K = (B'*X)*E
Ks = [K]
sizehint!(Ks, len)
observe_gdre_step!(observer, tstops[1], X, K)
# Global parameter for the method
CᵀC = C'C
for i in 2:len
τ = tstops[i-1] - tstops[i]
gF = (τ*(A-B*K)-E)/2
Fs, Es, Q, Z = schur(gF, Ed)
# Solve Lyapunov equation of 1st stage
AXE = A'X*E
R = CᵀC + AXE + AXE' - K'K
R = real(R+R')/2
utqu!(R, Z) # R = Z'*R*Z
lyapcs!(Fs, Es, R; adj=true)
K1 = R
utqu!(K1, Q') # K1 = Q*K1*Q'
# Solve Lyapunov equation of 2nd stage
EK1E = E'*K1*E
EK1B = E'*(K1*B)
R2 = -τ^2*(EK1B*EK1B')-2*EK1E
R2 = real(R2+R2')/2
utqu!(R2, Z) # R2 = Z'*R2*Z
lyapcs!(Fs, Es, R2; adj=true)
K21 = R2
utqu!(K21, Q') # K21 = Q*K21*Q'
K2 = K21 - K1
# Solve Lyapunov equation of 3rd stage
α = (24/25)*τ
β = (3/25)*τ
EK2E = E'*K2*E
EK2B = E'*(K2*B)
TMP = EK2B*EK1B'
R3 = (245/25)*EK1E + (36/25)*EK2E - (426/625)*τ^2*(EK1B*EK1B') - β^2*(EK2B*EK2B') - α*β*(TMP+TMP')
R3 = real(R3+R3')/2
utqu!(R3, Z) # R3 = Z'*R3*Z
lyapcs!(Fs, Es, R3; adj=true)
K31 = R3
utqu!(K31, Q') # K31 = Q*K31*Q'
K3 = K31 - (17/25)*K1
# Solve Lyapunov equation of 4th stage
R4 = -(981/125)*EK1E-(177/125)*EK2E-(1/5)*E'*K3*E
R4 = real(R4+R4')/2
utqu!(R4, Z) # R4 = Z'*R4*Z
lyapcs!(Fs, Es, R4; adj=true)
K41 = R4
utqu!(K41, Q') # K41 = Q*K41*Q'
K4 = K41 + K3
# Update X
X = X + τ*((19/18)*K1 + 0.25*K2 + (25/216)*K3 + (125/216)*K4)
save_state && push!(Xs, X)
# Update K
K = (B'*X)*E
push!(Ks, K)
observe_gdre_step!(observer, tstops[i], X, K)
end
save_state || push!(Xs, X)
observe_gdre_done!(observer)
return DRESolution(Xs, Ks, tstops)
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 1641 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
function _solve(
prob::GDREProblem{LDLᵀ{TL,TD}},
::Ros1;
dt::Real,
save_state::Bool,
adi_initprev::Bool=true,
adi_kwargs=NamedTuple(),
observer,
) where {TL,TD}
@timeit_debug "callbacks" observe_gdre_start!(observer, prob, Ros1())
T = LDLᵀ{TL,TD}
@unpack E, A, B, C, tspan = prob
q = size(C, 1)
X = prob.X0::T
tstops = tspan[1]:dt:tspan[2]
len = length(tstops)
# Output Trajectories
Xs = [X]
save_state && sizehint!(Xs, len)
L, D = X
BᵀLD = (B'*L)*D
K = BᵀLD*(L'*E)
Ks = [K]
sizehint!(Ks, len)
@timeit_debug "callbacks" observe_gdre_step!(observer, tstops[1], X, K)
for i in 2:len
τ = tstops[i-1] - tstops[i]
# Coefficient Matrix of the Lyapunov Equation
F = lr_update(A - E/(2τ), -1, B, K)
# Right-hand side:
G::TL = _hcat(TL, C', E'L)
S::TD = _dcat(TD, I(q), (BᵀLD)' * BᵀLD + D/τ)
R::T = compress!(LDLᵀ(G, S))
# Update X
lyap = GALEProblem(E, F, R)
initial_guess = adi_initprev ? X : nothing
X = @timeit_debug "ADI" solve(lyap, ADI(); observer, initial_guess, adi_kwargs...)
save_state && push!(Xs, X)
# Update K
L, D = X
BᵀLD = (B'*L)*D
K = BᵀLD*(L'*E)
push!(Ks, K)
@timeit_debug "callbacks" observe_gdre_step!(observer, tstops[i], X, K)
end
save_state || push!(Xs, X)
@timeit_debug "callbacks" observe_gdre_done!(observer)
return DRESolution(Xs, Ks, tstops)
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 2012 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
function _solve(
prob::GDREProblem{LDLᵀ{TL,TD}},
alg::Ros2;
dt::Real,
save_state::Bool,
observer,
adi_kwargs=NamedTuple(),
) where {TL,TD}
observe_gdre_start!(observer, prob, Ros2())
@unpack E, A, B, C, tspan = prob
q = size(C, 1)
X = prob.X0
tstops = tspan[1]:dt:tspan[2]
len = length(tstops)
# Global parameter for the method
γ = 1+(1/sqrt(2))
# Output Trajectories
Xs = [X]
save_state && sizehint!(Xs, len)
L, D = X
BᵀLD = (B'*L)*D
K = BᵀLD*(L'*E)
Ks = [K]
sizehint!(Ks, len)
observe_gdre_step!(observer, tstops[1], X, K)
for i in 2:len
τ = tstops[i-1] - tstops[i]
γτ = γ*τ
F = lr_update(γτ*A - E/2, inv(-γτ), B, K)
# Solve Lyapunov equation of 1st stage
G::TL = _hcat(TL, C', A'L, E'L)
n_G = size(G, 2)
n_L = size(L, 2)
S::TD = _zeros(TD, n_G)
b1 = 1:q
b2 = q+1:q+n_L
b3 = n_G-n_L+1:n_G
S[b1, b1] = I(q)
S[b2, b3] = D
S[b3, b2] = D
S[b3, b3] = - (BᵀLD)' * BᵀLD
R1 = compress!(LDLᵀ{TL,TD}(G, S))
lyap = GALEProblem(E, F, R1)
K1 = solve(lyap, ADI(); observer, adi_kwargs...)
# Solve Lyapunov equation of 2nd stage
T₁, D₁ = K1
BᵀT₁D₁ = (B'*T₁)*D₁
G₂::TL = E'T₁
S₂::TD = (τ^2 * BᵀT₁D₁)' * BᵀT₁D₁ + (2-1/γ) * D₁
R2 = LDLᵀ{TL,TD}(G₂, S₂)
lyap = GALEProblem(E, F, R2)
K2 = solve(lyap, ADI(); observer, adi_kwargs...)
# Update X
X = X + ((2-1/2γ)*τ)*K1 + (-τ/2)*K2
save_state && push!(Xs, X)
# Update K
L, D = X
BᵀLD = (B'*L)*D
K = BᵀLD*(L'*E)
push!(Ks, K)
observe_gdre_step!(observer, tstops[i], X, K)
end
save_state || push!(Xs, X)
observe_gdre_done!(observer)
return DRESolution(Xs, Ks, tstops)
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 5891 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
# Keep defaults in sync with docstring of NewtonADI!
function CommonSolve.solve(
prob::GAREProblem{TG,TQ},
::NewtonADI;
reltol = size(prob.A, 1) * eps(),
maxiters = 5,
observer = nothing,
adi_initprev::Bool = false,
adi_kwargs = NamedTuple(),
inexact::Bool = true,
inexact_hybrid::Bool = true,
inexact_forcing = quadratic_forcing,
linesearch::Bool = true,
) where {TG,TQ}
TG <: LDLᵀ{<:AbstractMatrix,UniformScaling{Bool}} || error("TG=$TG not yet implemented")
TQ <: LDLᵀ{<:AbstractMatrix,UniformScaling{Bool}} || error("TQ=$TQ not yet implemented")
@timeit_debug "callbacks" observe_gare_start!(observer, prob, NewtonADI())
TL = TD = Matrix{Float64}
@unpack E, A, Q = prob
B, _ = prob.G
Cᵀ, _ = Q
n = size(A, 2)
X = LDLᵀ{TL,TD}(zeros(n, 0), zeros(0, 0)) # this is ugly
res = Q
res_norm = norm(res)
abstol = reltol * res_norm
i = 0
local X_prev
while true
# Compute residual
L, D = X
EᵀL = E'L
BᵀLD = (B'L)*D
DLᵀGLD = (BᵀLD)'BᵀLD
K = BᵀLD * (EᵀL)'
res = residual(prob, X; EᵀL, DLᵀGLD)
res_norm_prev = res_norm
res_norm = norm(res)
if i > 0 && linesearch
@timeit_debug "Armijo line search" begin
α = 0.1
# The line search is mostly triggered for early Newton iterations `i`,
# where the linear systems to be solved have few columns and `X` has low rank.
# Therefore, an efficient implementation is not that important for now.
if res_norm > (1-α) * res_norm_prev
X̃ = X # backup if line search fails
β = 1/2 # Armijo parameter
λ = β # step size
while true
# Check sufficient decrease condition:
# (naive implementation)
X = (1 - λ) * X_prev + λ * X̃
res = residual(prob, X)
res_norm = norm(res)
if res_norm < (1 - λ*α) * res_norm_prev
@debug "Accepting line search λ=$λ"
# Update feedback matrix K and other auxillary variables:
# (naive implementation)
L, D = X
EᵀL = E'L
BᵀLD = (B'L)*D
DLᵀGLD = (BᵀLD)'BᵀLD
K .= BᵀLD * (EᵀL)'
break
end
# Prepare next step size:
λ *= β
if λ < eps()
@warn "Line search failed; using un-modified iterate"
λ = 1.0
X = X̃
break
end
end
@timeit_debug "callbacks" observe_gare_metadata!(observer, "line search", λ)
end
end
end
@timeit_debug "callbacks" observe_gare_step!(observer, i, X, res, res_norm)
res_norm <= abstol && break
if i >= maxiters
@timeit_debug "callbacks" observe_gare_failed!(observer)
@warn "NewtonADI did not converge" residual=res_norm abstol maxiters
break
end
i += 1
# Coefficient Matrix of the Lyapunov Equation
F = lr_update(A, -1, B, K)
# Right-hand side:
m = size(B, 2)
q = size(Cᵀ, 2)
EᵀXB = EᵀL * (BᵀLD)'
G::TL = _hcat(TL, Cᵀ, EᵀXB)
S::TD = _dcat(TD, I(q), I(m))
RHS = LDLᵀ(G, S)
# ADI setup
lyap = GALEProblem(E, F, RHS)
initial_guess = adi_initprev ? X : nothing
adi_reltol = get(adi_kwargs, :reltol, reltol / 10)
if inexact
η = inexact_forcing(i, res_norm)
adi_abstol = η * res_norm
if inexact_hybrid
# If the classical/"exact" tolerance is less strict than
# the one of the Inexact Newton, use that tolerance instead.
classical_abstol = adi_reltol * norm(lyap.C)
switch_back = classical_abstol > adi_abstol
@timeit_debug "callbacks" observe_gare_metadata!(observer, "inexact", !switch_back)
if switch_back
@debug "Switching from inexact to classical Newton method" i inexact_abstol=adi_abstol classical_abstol
adi_abstol = classical_abstol
end
else
@timeit_debug "callbacks" observe_gare_metadata!(observer, "inexact", true)
end
else
adi_abstol = adi_reltol * norm(lyap.C)
end
# Newton step:
X_prev = X
X = @timeit_debug "ADI" solve(
lyap, ADI();
maxiters=100,
observer,
initial_guess,
abstol=adi_abstol,
adi_kwargs...)
end
@timeit_debug "callbacks" observe_gare_done!(observer, i, X, res, res_norm)
X
end
"""
superlinear_forcing(i, _) = 1 / (i^3 + 1)
Exemplary forcing term to obtain superlinear convergence in the inexact Newton method.
`i::Int` refers to the current Newton step.
See [`NewtonADI`](@ref).
"""
superlinear_forcing(i, _) = 1 / (i^3 + 1)
"""
quadratic_forcing(_, residual_norm) = min(0.1, 0.9 * residual_norm)
Exemplary forcing term to obtain quadratic convergence in the inexact Newton method.
`residual_norm::Float64` refers to the norm of the previous Newton residual.
See [`NewtonADI`](@ref).
"""
quadratic_forcing(_, residual_norm) = min(0.1, 0.9 * residual_norm)
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 960 |
# This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
using Compat: @something
@timeit_debug "residual(::GAREProblem, ::LDLᵀ)" function residual(
prob::GAREProblem,
X::LDLᵀ;
AᵀL = nothing,
EᵀL = nothing,
BᵀLD = nothing,
DLᵀGLD = nothing,
)
@unpack E, A, Q, G = prob
Cᵀ, _ = Q
B, _ = G
L, D = X
h = size(Cᵀ, 2)
zₖ = size(L, 2)
dim = h + 2zₖ
@debug "Assembling ARE residual" h zₖ
# Compute optional inputs
AᵀL = @something(AᵀL, A'L)
EᵀL = @something(EᵀL, E'L)
if DLᵀGLD === nothing
BᵀLD = @something(BᵀLD, (B'L)*D)
DLᵀGLD = (BᵀLD)'BᵀLD
end
# Compute residual following Benner, Li, Penzl (2008)
R = [Cᵀ AᵀL EᵀL]
T = zeros(dim, dim)
b1 = 1:h
b2 = h+1:h+zₖ
b3 = b2 .+ zₖ
for i in b1
T[i, i] = 1
end
T[b2, b3] .= T[b3, b2] .= D
T[b3, b3] .= -DLᵀGLD
LDLᵀ(R, T)
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 3389 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
"""
Generalized differential Riccati equation
E'ẊE = C'C + A'XE + E'XA - E'XBB'XE
X(t0) = X0
having the fields `E`, `A`, `C`, `X0`, and `tspan`=`(t0, tf)`.
"""
struct GDREProblem{XT}
E
A
B
C
X0::XT
tspan
GDREProblem(E, A, B, C, X0::XT, tspan) where {XT} = new{XT}(E, A, B, C, X0, tspan)
end
"""
Solution to a generalized differential Riccati equation (DRE)
as returned by [`solve(::GDREProblem, alg; kwargs...)`](@ref GDREProblem).
The solution has three fields:
* `X::Vector{T}`: state `X(t)`; `T` may be a `Matrix` or [`LDLᵀ`](@ref)
* `K::Vector{<:Matrix}`: feedback `K(t) := B' * X(t) * E`
* `t::Vector{<:Real}`: discretization time
By default, the state `X` is only stored at the boundaries of the time span,
as one is mostly interested only in the feedback matrices `K`.
To store the full state trajectory, pass `save_state=true` to `solve`.
"""
struct DRESolution
X
K
t
end
"""
Generalized algebraic (continuous time) algebraic Riccati equation
Q + A'XE + E'XA - E'XGXE = 0
"""
struct GAREProblem{TG,TQ}
E
A
G::TG
Q::TQ
end
abstract type AlgebraicRiccatiSolver end
"""
NewtonADI()
Kleinman-Newton method to solve algebraic Riccati equations.
The algebraic Lyapunov equations arizing at every Newton steps are solved using the [`ADI`](@ref).
solve(prob::GAREProblem, NewtonADI(); kwargs...)
Supported keyword arguments:
* `reltol = size(prob.A, 1) * eps()`: relative Riccati residual tolerance
* `maxiters = 5`: maximum number of Newton steps
* `observer`: see [`Callbacks`](@ref)
* `adi_initprev = false`: whether to use previous Newton iterate
as the initial guess for the [`ADI`](@ref).
If `false`, the default initial value of zero is used.
* `adi_kwargs::NamedTuple`:
keyword arguments to pass to `solve(_, ::ADI; adi_kwargs...)`
* `inexact = true`:
whether to allow (more) inexact Lyapunov solutions
* `inexact_forcing = quadratic_forcing`:
compute the forcing parameter `η = inexact_forcing(i, residual_norm)`
as described by Dembo et al. (1982), where
`i::Int` is the Newton step and
`residual_norm::Float64` is the norm of the Riccati residual.
See [`quadratic_forcing`](@ref), and [`superlinear_forcing`](@ref).
* `inexact_hybrid = true`:
whether to switch to the classical Newton method,
if the absolute Lyapunov tolerance of the classical Newton method
is less strict (i.e. larger) than the tolerance `η * residual_norm`.
* `linesearch = true`: whether to perform an Armijo line search
if the Riccati residual did not decrease sufficiently,
see e.g. Benner et al. (2015).
Default arguments to Lyapunov solver, which can all be overwritten by `adi_kwargs`:
* `maxiters = 100`: maximum number of ADI iterations
* `observer = observer`
* `initial_guess`: see `adi_initprev` above
* `reltol`: defaults a fraction of the Riccati tolerance, `reltol/10`
* `abstol`: controlled by `inexact*` above, if `inexact = true`.
References:
* Dembo, Eisenstat, Steihaug: Inexact Newton Methods. 1982.
https://doi.org/10.1137/0719025
* Benner, Heinkenschloss, Saak, Weichelt: Inexact low-rank Newton-ADI method for large-scale algebraic Riccati equations. 2015.
http://www.mpi-magdeburg.mpg.de/preprints/
"""
struct NewtonADI <: AlgebraicRiccatiSolver end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 2690 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
using Base.Iterators: Stateful, cycle
"""
Cyclic(::Shifts.Strategy)
Cyclic(values)
Cycle through precomputed `values` or the shifts produced by the inner strategy.
That is, continue with the first parameter once the last one has been consumed.
Examples:
```julia
Cyclic(Heuristic(10, 20, 20))
Cyclic(Float64[-1, -2, -3])
```
"""
struct Cyclic <: Strategy
inner
end
"""
Wrapped(func!, ::Shifts.Strategy)
Apply `func!` to the set of shifts produced by the inner strategy via [`Shifts.take_many!`](@ref).
This strategy may be used, e.g., to filter or reorder the shifts.
Complex-valued shifts must occur in conjugated pairs.
Examples:
```julia
Wrapped(reverse, Projection(2))
Wrapped(Projection(4)) do shifts
filter(s -> real(s) < -1, shifts)
end
```
"""
struct Wrapped <: Strategy
func!
inner::Strategy
end
###
"""
BufferedIterator(generator)
Initialize an internal buffer of type `Vector{<:Number}` from
[`Shifts.take_many!(generator)`](@ref Shifts.take_many!)
and return shifts one-by-one using `popfirst!`.
Refill the buffer once it is depleated.
"""
mutable struct BufferedIterator
buffer::Vector{<:Number}
generator
BufferedIterator(gen) = new(ComplexF64[], gen)
end
"""
Shifts.take_many!(generator)
Return a `Vector{<:Number}` of shift parameters to be used
within a [`Shifts.BufferedIterator`](@ref).
"""
take_many!
mutable struct WrappedIterator
func!
generator
end
# Allow Cyclic(42) for convenience:
_init(values, _) = values
_init(s::Strategy, prob) = init(s, prob)
init(c::Cyclic, prob) = Stateful(cycle(_init(c.inner, prob)))
# Ensure that BufferedIterator remains the outer-most structure:
_wrap(it, func!) = WrappedIterator(func!, it)
_wrap(it::BufferedIterator, func!) = BufferedIterator(WrappedIterator(func!, it.generator))
init(w::Wrapped, prob) = _wrap(init(w.inner, prob), w.func!)
update!(it::BufferedIterator, args...) = update!(it.generator, args...)
update!(it::WrappedIterator, args...) = update!(it.generator, args...)
take_many!(it::WrappedIterator) = it.func!(take_many!(it.generator))
function take!(it::BufferedIterator)
if isempty(it.buffer)
it.buffer = take_many!(it.generator)
@debug "Obtained $(length(it.buffer)) new shifts"
end
# TODO: Using `popfirst!` feels inefficient, even though there should be only 10s of elements buffered.
popfirst!(it.buffer)
end
###
"""
safe_sort!(shifts)
Ensure that complex conjugated values are located adjacent to one another.
"""
safe_sort!(shifts) = sort!(shifts; by = v -> (real(v), abs(imag(v))))
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 2307 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
using UnPack
"""
Shifts.Heuristic(nshifts, k₊, k₋)
Compute heuristic or sub-optimal shift parameters following Algorithm 5.1 of
> Penzl: A cyclic low rank Smith method for large sparse Lyapunov equations,
> SIAM J. Sci. Comput., 21 (1999), pp. 1401-1418. DOI: 10.1137/S1064827598347666
"""
struct Heuristic <: Strategy
nshifts::Int
k₊::Int
k₋::Int
end
function init(strategy::Heuristic, prob)
@unpack nshifts, k₊, k₋ = strategy
@unpack E, A = prob
# TODO: Make solver configurable.
# TODO: Think about caching of properties of E.
# The matrix E shouldn't change all that much between iterations of the same algorithm,
# or between algorithms in general.
Ef = factorize(E)
b0 = ones(size(E, 1))
R₊ = compute_ritz_values(x -> Ef \ (A * x), b0, k₊, "E⁻¹A")
R₋ = compute_ritz_values(x -> A \ (E * x), b0, k₋, "A⁻¹E")
# TODO: R₊ and R₋ may not be disjoint. Remove duplicates, or replace values that differ
# by an eps with their average.
R = vcat(R₊, inv.(R₋))
heuristic(R, nshifts)
end
function heuristic(R, nshifts=length(R))
s(t, P) = prod(abs(t - p) / abs(t + p) for p in P)
p = argmin(R) do p
maximum(s(t, (p,)) for t in R)
end
P = isreal(p) ? [p] : [p, conj(p)]
while length(P) < nshifts
p = argmax(R) do t
s(t, P)
end
if isreal(p)
push!(P, p)
else
append!(P, (p, conj(p)))
end
end
return P
end
function compute_ritz_values(A, b0, k::Int, desc::String)
n = length(b0)
H = zeros(k + 1, k)
V = zeros(n, k + 1)
V[:, 1] .= (1.0 / norm(b0)) * b0
# Arnoldi
for j in 1:k
w = A(V[:, j])
# Repeated modified Gram-Schmidt (MGS)
for _ = 1:2
for i = 1:j
g = V[:, i]' * w
H[i, j] += g
w -= V[:, i] * g
end
end
H[j+1, j] = beta = norm(w)
V[:, j+1] .= (1.0 / beta) * w
end
ritz = eigvals(@view H[1:k, 1:k])
isstable(v) = real(v) < 0
all(isstable, ritz) && return ritz
@warn "Discarding unstable Ritz values of $desc"
filter!(isstable, ritz)
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 1878 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
using LinearAlgebra, SparseArrays
using UnPack
using Compat: keepat!
"""
Shifts.Projection(u::Int)
Compute shift parameters based on the `u` most recent increments comprising the solution candidate.
Only even `u > 1` are allowed, such that an ADI double-step can properly be accounted for.
See section 5.3.1 of
> Kürschner: Efficient low-rank solution of large-scale matrix equations.
> Otto-von-Guericke-Universität Magdeburg (2016).
The strategy has first been presented in
> Benner, Kürschner, Saak: Self-generating and efficient shift parameters in ADI methods for large Lyapunov and Sylvester equations,
> Electronic Transactions on Numerical Analysis, 43 (2014), pp. 142-162.
> https://etna.math.kent.edu/volumes/2011-2020/vol43/abstract.php?vol=43&pages=142-162
"""
struct Projection <: Strategy
n_history::Int
function Projection(u)
isodd(u) && throw(ArgumentError("History must be even; got $u"))
new(u)
end
end
mutable struct ProjectionShiftIterator
prob
n_history::Int
Vs::Vector{Any}
end
function init(strategy::Projection, prob)
it = ProjectionShiftIterator(prob, strategy.n_history, [])
BufferedIterator(it)
end
function update!(it::ProjectionShiftIterator, _, R, Vs...)
isempty(Vs) && push!(it.Vs, R)
append!(it.Vs, Vs)
lst = length(it.Vs)
fst = max(1, lst - it.n_history + 1)
keepat!(it.Vs, fst:lst)
return
end
function take_many!(it::ProjectionShiftIterator)
@unpack E, A = it.prob
@unpack Vs = it
N = hcat(Vs...)::AbstractMatrix{<:Real}
Q = orth(N)
Ẽ = restrict(E, Q)
à = restrict(A, Q)
λ = eigvals(Ã, Ẽ)
# TODO: flip values at imaginary axes instead
λ₋ = filter(l -> real(l) < 0, λ)
safe_sort!(λ₋)
return λ₋
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 421 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
_dcat(::Type{T}, X, Xs...) where {T} = _dcat(T, (X, Xs...))
function _dcat(::Type{T}, Xs) where {T}
n = sum(X -> size(X, 1), Xs)
D = _zeros(T, n, n)
k = 0
for X in Xs
l = size(X, 1)
span = k+1:k+l
D[span,span] = X
k += l
end
@assert k == n
return D
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 196 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
_diagm(::Type{<:Matrix}, v) = diagm(v)
_diagm(::Type{<:SparseMatrixCSC}, v) = spdiagm(v)
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 445 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
_hcat(::Type{T}, X, Xs...) where {T} = _hcat(T, (X, Xs...))
function _hcat(::Type{T}, Xs) where {T}
m = size(first(Xs), 1)
n = sum(X -> size(X, 2), Xs)
L = _zeros(T, m, n)
k = 0
for X in Xs
l = size(X, 2)
span = k+1:k+l
L[:,span] = X
k += l
end
@assert k == n
return L
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 275 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
_zeros(::Type{Matrix{T}}, m::Int, n::Int=m) where {T} = zeros(T, m, n)
_zeros(::Type{<:SparseArrays.SparseMatrixCSC{T}}, m::Int, n::Int=m) where {T} = spzeros(T, m, n)
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 244 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
import .Stuff: restrict
function restrict(AUV::LowRankUpdate, Q)
A, α, U, V = AUV
restrict(A, Q) + inv(α) * ((Q'U) * (V*Q))
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 3032 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
using Test, DifferentialRiccatiEquations
using LinearAlgebra
n = 10
k = 2
@testset "Conversions" begin
L = randn(n, k)
D = rand(k, k)
X = LDLᵀ(L, D)
M = Matrix(X)
@test M isa Matrix{Float64}
@test size(M) == (n, n)
@test M ≈ L*D*L'
@test norm(M) ≈ norm(X)
end
function sample(n, k)
local D, L
while true
L = randn(n, k)
rank(L) == k && break
end
λ = rand(k) .+ 0.1
D = diagm(λ)
return LDLᵀ(L, D)
end
@testset "Rank k" begin
X = sample(n, k)
M = Matrix(X)
@test M isa Matrix{Float64}
@test rank(X) == rank(M) == k
_L = only(X.Ls)
_D = only(X.Ds)
L, D = X
@test L === _L
@test D === _D
for d in 2:k
D[d,d] = 0
end
compress!(X)
@test rank(X) == 1
end
@testset "Rank 0" begin
X = LDLᵀ(randn(n, 1), zeros(1, 1))
@test rank(X) == 1 # not the actual rank
compress!(X)
@test rank(X) == 0
@test Matrix(X) == zeros(n, n)
X = sample(n, 0)
@test rank(X) == 0
@test Matrix(X) == zeros(n, n)
Z = zero(X)
@test typeof(Z) == typeof(X)
@test rank(Z) == 0
end
@testset "Compression" begin
# TODO: Once compression is configurable, this must be adjusted.
@assert 2k < 0.5n # U+U does not trigger compression
U = sample(n, k)
V = Matrix(U)
W = U + U
@test rank(V) == k
@test rank(W) == 2k
@test Matrix(W) ≈ 2V
@testset "Implicit Compression" begin
# Implicit compression upon iteration:
W = U + U
@test rank(W) == 2k
L, D = W
@test rank(W) == k
@test size(L, 1) == n
@test size(L, 2) == size(D, 1) == size(D, 2) == k
@test Matrix(W) ≈ L*D*L' ≈ 2V
# Repeated iteration does not alter the components:
L1, D1 = W
@test L1 === L
@test D1 === D
end
@testset "Skipped Compression" begin
# Don't compress singleton components:
W = U + U
concatenate!(W)
@test rank(W) == 2k
L, D = W
@test size(L, 1) == n
@test size(L, 2) == size(D, 1) == size(D, 2) == 2k
@test Matrix(W) ≈ L*D*L' ≈ 2V
end
desc = ("w/ ", "w/o")
concat = (true, false)
@testset "Explicit Compression $d Concatenation" for (d, cc) in zip(desc, concat)
# Explicit compression reduces rank:
W = U + U
cc && concatenate!(W)
@test rank(W) == 2k
compress!(W)
@test rank(W) == k
L, D = W
@test size(L, 1) == n
@test size(L, 2) == size(D, 1) == size(D, 2) == k
@test Matrix(W) ≈ L*D*L' ≈ 2V
end
end
@testset "Arithmetic" begin
# TODO: Once compression is configurable, this must be adjusted.
@assert 3k > 0.5n # X+X+X does trigger compression
X = sample(n, k)
@test rank(X) == k
@test rank(compress!(X+X)) == k
@test rank(X+X+X) == k
#@test rank(X-X) == 0 # flaky
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 1453 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
using Test
using LinearAlgebra
using DifferentialRiccatiEquations: LowRankUpdate, lr_update
using SparseArrays
n = 10
k = 3
@testset "Dense A" begin
A = rand(n, n)
U = rand(n, k)
V = rand(k, n)
AUV = lr_update(A, 1, U, V)
@test AUV isa Matrix
end
function test_lr_update(A, U, V)
AUV = lr_update(A, -1, U, V) # technically a downdate
@test AUV isa LowRankUpdate
# Decomposition:
_A, _α, _U, _V = AUV
@test _A === A
@test _U === U
@test _V === V
@test _α === -1
# Vector solve:
M = A - U*V
B = rand(n)
X = AUV \ B
@test M * X ≈ B
# Matrix solve:
B1 = rand(n, 1)
X1 = AUV \ B1
@test M * X1 ≈ B1
# Addition:
E = sprand(n, n, 0.2)
EUV = AUV + E
@test typeof(EUV) == typeof(AUV)
_E, _α, _U, _V = EUV
@test _E ≈ A + E
@test _U === U
@test _V === V
@test _α === -1
end
@testset "Sparse A" begin
A = spdiagm(1.0:n) # invertible
A[2,1] = 1 # not diagonal
A[3,2] = 2 # not triangular or symmetric
desc = ["Dense", "Sparse"]
Us = [rand(n, k), sprand(n, k, 0.5)]
Vs = [rand(k, n), sprand(k, n, 0.5)]
tests = Iterators.product(
zip(desc, Us),
zip(desc, Vs),
)
@testset "$descU U, $descV V" for ((descU, U), (descV, V)) in tests
test_lr_update(A, U, V)
end
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 5171 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
using LinearAlgebra
using SparseArrays
using Test
using DifferentialRiccatiEquations
using DifferentialRiccatiEquations: Shifts
using .Shifts
using .Shifts: take!, init
penzl(p) = [-1 p; -p -1]
modified_penzl(v) = abs(real(v)) * penzl(imag(v) / real(v))
n = 3
E = sparse(1.0I(n))
A = spzeros(n, n)
A[1:2, 1:2] = penzl(1)
A[3:n, 3:n] .= -1/2
# Internally, the Ritz values are computed with a naive Arnoldi implementation,
# which is not very accurate. Therefore, the following testset is mostly broken.
# However, in practise, the shifts it produces work much better than accurate Ritz values.
@testset "Heuristic Penzl Shifts" begin
k = 2
strategy = Heuristic(k, 2, 2)
shifts = init(strategy, (; E, A))
@test shifts isa Vector{ComplexF64}
@test k <= length(shifts) <= k + 1
@test_broken any(≈(-1 + im), shifts)
@test_broken any(≈(-1 - im), shifts)
if length(shifts) > k
# The strategy may only report more shifts than requested
# if the last one has been complex, i.e., both the complex
# parameters as well as its conjugate had to be returned.
# In that case, the real shift had to be the first one.
@test any(≈(-1/2), shifts)
@test shifts[1] ≈ -1/2
end
# Ensure complex shifts occur in conjugated pairs:
i = findfirst(!isreal, shifts)
@test_broken !isnothing(i)
@test_broken shifts[i] == conj(shifts[i+1])
end
@testset "Cyclic helper" begin
shifts = init(Cyclic(1:3), nothing)
@test take!(shifts) == 1
@test take!(shifts) == 2
@test take!(shifts) == 3
@test take!(shifts) == 1
@testset "Type Stability ($(eltype(values)))" for values in (
1:2, # iterable
(1.0, 2.0), # Tuple
ComplexF64[1, 2], # Vector
)
shifts = init(Cyclic(values), nothing)
a, b = values
# Check value and type:
@test take!(shifts) === a
@test take!(shifts) === b
end
shifts = init(Cyclic(Heuristic(1, 1, 1)), (; E, A))
p = take!(shifts)
if isreal(p)
@test take!(shifts) == p
else
@test take!(shifts) == conj(p)
@test take!(shifts) == p
end
end
struct Dummy <: Shifts.Strategy values end
struct DummyIterator values end
Shifts.init(d::Dummy, _) = Shifts.BufferedIterator(DummyIterator(d.values))
Shifts.take_many!(d::DummyIterator) = d.values
# Prerequisite:
@test init(Dummy(nothing), nothing) isa Shifts.BufferedIterator
@testset "BufferedIterator helper" begin
@testset "Type Stability $(eltype(values))" for values in (
[1, 2, 3],
ComplexF64[1, 2, 3],
)
shifts = init(Dummy(copy(values)), nothing)
# Check value and type stability:
for v in values
@test take!(shifts) === v
end
end
end
@testset "Wrapped helper" begin
shifts = init(Wrapped(reverse, Dummy([1,2,3])), nothing)
@test shifts isa Shifts.BufferedIterator
@test shifts.generator isa Shifts.WrappedIterator
@test shifts.generator.generator isa DummyIterator
@test take!(shifts) == 3
@test take!(shifts) == 2
@test take!(shifts) == 1
end
@testset "Adaptive Projection Shifts" begin
@test_throws ArgumentError Projection(1)
strategy = Projection(2)
shifts = init(strategy, (; E, A))
# Ensure that no shifts have been computed so far:
@test shifts isa Shifts.BufferedIterator
@test isempty(shifts.buffer)
# Pass some initial data:
Shifts.update!(shifts, LDLᵀ(zeros(n, 0), zeros(0, 0)), ones(n))
@test isempty(shifts.buffer)
# As the initial residual was rank one,
# only one shift should have been computed:
@test Shifts.take!(shifts) ≈ -5/6
@test isempty(shifts.buffer)
end
function preserves_conj_pairs(shifts, n=length(shifts); verbose=true)
i = 0
while i < n
i += 1
v = take!(shifts)
if !isreal(v)
i += 1
w = take!(shifts)
w ≈ conj(v) && continue
verbose && @error "Error at shift $i: expected conj($v), got $w"
return false
end
end
return true
end
# Ensure that complex shifts occur in conjugated pairs.
@testset "Conjugated Pairs" begin
@testset "Same $desc" for (desc, f) in [
("magnitude", a -> -exp(a*im)),
("real part", a -> -1 - a*im),
]
@testset "Helper $(length(vals))" for vals in (-1:1, -3:2:3)
vals = [f(v) for v in -n:2:n]
@test !preserves_conj_pairs(copy(vals); verbose=false)
Shifts.safe_sort!(vals)
@test preserves_conj_pairs(copy(vals))
end
@testset "Hacky Projection shifts" begin
I4 = 1.0 * I(4)
A = zeros(4, 4)
A[1:2, 1:2] .= modified_penzl(f(1))
A[3:4, 3:4] .= modified_penzl(f(2))
shifts = init(Shifts.Projection(2), (; E=I4, A))
# Hack input such that full spectrum of A is returned.
Shifts.update!(shifts, nothing, nothing, I4)
@test preserves_conj_pairs(shifts, 4)
end
end
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 2909 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
using Test
using DifferentialRiccatiEquations
using LinearAlgebra, SparseArrays
using MAT, UnPack
const DREs = DifferentialRiccatiEquations
# Dense Setup
P = matread(joinpath(@__DIR__, "Rail371.mat"))
@unpack E, A, B, C, X0 = P
tspan = (4500., 4400.) # backwards in time
prob = GDREProblem(E, A, B, C, X0, tspan)
# Low-Rank Setup With Dense D
q = size(C, 1)
L = E \ C'
D = Matrix(0.01I(q))
X0s = LDLᵀ(L, D)
sprob1 = GDREProblem(E, A, B, C, X0s, tspan)
# Low-Rank Setup With Sparse D
Ds = sparse(0.01I(q))
X0ss = LDLᵀ(L, Ds)
sprob2 = GDREProblem(E, A, B, C, X0ss, tspan)
Δt(nsteps::Int) = (tspan[2] - tspan[1]) ÷ nsteps
function smoketest(prob, alg)
sol = solve(prob, alg; dt=Δt(1))
@test sol isa DREs.DRESolution
@test length(sol.X) == 2 # only store first and last state by default
@test first(sol.X) === prob.X0 # do not copy
sol = solve(prob, alg; dt=Δt(2), save_state=true)
@test sol isa DREs.DRESolution
@test length(sol.t) == length(sol.X) == length(sol.K) == 3
@test issorted(sol.t) == issorted(tspan) # do not alter direction of time
end
@testset "Dense $alg" for alg in (Ros1(), Ros2(), Ros3(), Ros4())
smoketest(prob, alg)
end
# Verify Low-Rank Setup
@test Matrix(X0s) ≈ X0
@test Matrix(X0ss) ≈ X0
@testset "Low-Rank Ros1()" begin
alg = Ros1()
# Replicate K with dense solver:
ref = solve(prob, alg; dt=Δt(5))
ε = norm(ref.K[end]) * size(E, 1) * eps() * 100
@testset "Dense D" begin
smoketest(sprob1, alg)
sol1 = solve(sprob1, alg; dt=Δt(5))
@test norm(ref.K[end] - sol1.K[end]) < ε
end
@testset "Sparse D" begin
smoketest(sprob2, alg)
sol2 = solve(sprob2, alg; dt=Δt(5))
@test norm(ref.K[end] - sol2.K[end]) < ε
end
end
@testset "Low-Rank Ros2()" begin
alg = Ros2()
# Replicate K with dense solver:
ref = solve(prob, alg; dt=Δt(5))
ε = norm(ref.K[end]) * size(E, 1) * eps() * 100
@testset "Dense D" begin
smoketest(sprob1, alg)
sol1 = solve(sprob1, alg; dt=Δt(5))
@test norm(ref.K[end] - sol1.K[end]) < ε
end
@testset "Sparse D" begin
smoketest(sprob2, alg)
sol2 = solve(sprob2, alg; dt=Δt(5))
@test norm(ref.K[end] - sol2.K[end]) < ε
end
end
using DifferentialRiccatiEquations: residual
using DifferentialRiccatiEquations.Shifts
@testset "NewtonADI()" begin
G = LDLᵀ(B, I)
Q = LDLᵀ(C', I)
are = GAREProblem(E, A, G, Q)
reltol = 1e-10
@testset "$(adi_kwargs.shifts)" for adi_kwargs in [
(shifts = Projection(2),), # leads to some complex shifts
(shifts = Cyclic(Heuristic(10, 20, 20)), maxiters = 200),
]
X = solve(are, NewtonADI(); reltol, adi_kwargs, maxiters=10)
@test norm(residual(are, X)) < reltol * norm(Q)
end
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | code | 695 | # This file is a part of DifferentialRiccatiEquations. License is MIT: https://spdx.org/licenses/MIT.html
using Test
using DifferentialRiccatiEquations
using SparseArrays
using DifferentialRiccatiEquations.Stuff: orth
@testset "DifferentialRiccatiEquations.jl" begin
@testset "LDLᵀ" begin include("LDLt.jl") end
@testset "LowRankUpdate" begin include("LowRankUpdate.jl") end
@testset "orth" begin
N = zeros(4, 1)
Q = orth(N)
@test size(Q) == (4, 0)
Ns = sparse(N)
Qs = orth(Ns)
@test size(Qs) == (4, 0)
end
@testset "ADI Shifts" begin include("Shifts.jl") end
@testset "Oberwolfach Rail" begin include("rail.jl") end
end
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | docs | 1625 | # v0.4
* Fix LDLᵀ compression for indefinite objects (d6a649ad62ab6d413ce82e5a2b0090de813a33de)
* Add callbacks to allow user to gather information during `solve` calls;
see docstring of the `Callbacks` module for more info
* Add configurable shift strategies;
see docstring of the `Shifts` module for more info
* Change default shift strategy.
While this is not API breaking, it does affect the convergence behavior.
(7b32660af73c23c0d710b215705688842aa0bb70)
* Fix order of automatic/projection shifts: ensure that complex shifts occur in
conjugated pairs directly one after the other (e36a1163f9db4796b334fbdf23c23ea4fd0aab9d)
* Add Inexact Newton method following Dembo et al. (1982) and Benner et al. (2015) to solve AREs;
see docstring of `NewtonADI` for more info
# v0.3
* Add license
* Improve documentation
* Breaking: Rename keyword arguments of `solve(::GALEProblem, ::ADI; nsteps, rtol)` to `maxiters` and `reltol`
* Rename default branch to `main`
# v0.2.2
* Fix ADI (425d4001112fcff88b30c58f020b106e10a7ef7b)
# v0.2.1
* Add low-rank Ros2 (1345647c610c4561e0d63e8fbee65a85693d8156)
# v0.2
* Add LDLᵀ factorization (4811939893a98b6ebc6e442f6a85ff0dcde4b42e)
* Add LowRankUpdate representation which supports `\` via Sherman-Morrison-Woodbury (2b00c7bf0d817973d41d883773a68db173faaaa6)
* Add low-rank Ros1 (implicit Euler) (c1d4bcf5c22fb71f85512e78c0071e58ffaf1397)
* Dense solvers now support sparse `E` (331094d0ca4cc84f4ae2d13df41cc52b5d229663)
# v0.1
* Port Rosenbrock solvers from Lang 2017
* Reuse Schur decomposition within Rosenbrock steps (3706742ac179c312b66de1ec41d57a7c2924a7af)
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.4.0 | 95f5d5624ccbbd507bff326d2a7eee5be5dbfc8a | docs | 6004 | # DifferentialRiccatiEquations.jl
[](https://gitlab.mpi-magdeburg.mpg.de/jschulze/DifferentialRiccatiEquations.jl/pipelines)
[](https://gitlab.mpi-magdeburg.mpg.de/jschulze/DifferentialRiccatiEquations.jl/commits/main)
This package provides algorithms to solve autonomous Generalized Differential Riccati Equations (GDRE)
```math
\left\{
\begin{aligned}
E^T \dot X E &= C^T C + A^T X E + E^T X A - E^T X BB^T X E,\\
X(t_0) &= X_0.
\end{aligned}
\right.
```
More specifically:
* Dense Rosenbrock methods of orders 1 to 4
* Low-rank symmetric indefinite (LRSIF) Rosenbrock methods of order 1 and 2, $X = LDL^T$
In the latter case, the (generalized) Lyapunov equations arizing in the Rosenbrock stages
are solved using a LRSIF formulation of the Alternating-Direction Implicit (ADI) method,
as described by [LangEtAl2015].
The ADI uses the self-generating parameters described by [Kuerschner2016].
> **Warning**
> The low-rank 2nd order Rosenbrock method suffers from the same problems as described by [LangEtAl2015].
[Kuerschner2016]: https://hdl.handle.net/11858/00-001M-0000-0029-CE18-2
[LangEtAl2015]: https://doi.org/10.1016/j.laa.2015.04.006
The user interface hooks into [CommonSolve.jl] by providing the `GDREProblem` problem type
as well as the `Ros1`, `Ros2`, `Ros3`, and `Ros4` solver types.
[CommonSolve.jl]: https://github.com/SciML/CommonSolve.jl
# Getting started
The package can be installed from Julia's REPL:
```
pkg> add git@gitlab.mpi-magdeburg.mpg.de:jschulze/DifferentialRiccatiEquations.jl.git
```
To run the following demos, you further need the following packages and standard libraries:
```
pkg> add LinearAlgebra MAT SparseArrays UnPack
```
What follows is a slightly more hands-on version of `test/rail.jl`.
Please refer to the latter for missing details.
## Dense formulation
The easiest setting is perhaps the dense one,
i.e. the system matrices `E`, `A`, `B`, and `C`
as well as the solution trajectory `X` are dense.
First, load the system matrices from e.g. `test/Rail371.mat`
(see [License](#license) section below)
and define the problem parameters.
```julia
using DifferentialRiccatiEquations
using LinearAlgebra
using MAT, UnPack
P = matread("Rail371.mat")
@unpack E, A, B, C, X0 = P
tspan = (4500., 0.) # backwards in time
```
Then, instantiate the GDRE and call `solve` on it.
```julia
prob = GDREProblem(E, A, B, C, X0, tspan)
sol = solve(prob, Ros1(); dt=-100)
```
The trajectories $X(t)$, $K(t) := B^T X(t) E$, and $t$ may be accessed as follows.
```julia
sol.X # X(t)
sol.K # K(t) := B^T X(t) E
sol.t # discretization points
```
By default, the state $X$ is only stored at the boundaries of the time span `tspan`,
as one is mostly interested only in the feedback matrices $K$.
To store the full state trajectory, pass `save_state=true` to `solve`.
```julia
sol_full = solve(prob, Ros1(); dt=-100, save_state=true)
```
## Low-rank formulation
Continuing from the dense setup,
assemble a low-rank variant of the initial value,
$X_0 = LDL^T$ where $E^T X_0 E = C^T C / 100$ in this case.
Both dense and sparse factors are allowed for $D$.
```julia
using SparseArrays
q = size(C, 1)
L = E \ C'
D = sparse(0.01I(q))
X0_lr = LDLᵀ(L, D)
Matrix(X0_lr) ≈ X0
```
Passing this low-rank initial value to the GDRE instance
selects the low-rank algorithms and computes the whole trajectories in $X$ that way.
Recall that these trajectories are only stored iff one passes the keyword argument `save_state=true` to `solve`.
```julia
prob_lr = GDREProblem(E, A, B, C, X0_lr, tspan)
sol_lr = solve(prob_lr, Ros1(); dt=-100)
```
> **Note**
> The type of the initial value, `X0` or `X0_lr`,
> dictates the type used for the whole trajectory, `sol.X` and `sol_lr.X`.
## Solver introspection / Callbacks
To record information during the solution process,
e.g. the residual norms of every ADI step at every GDRE time step,
define a custom observer object and associated callback methods.
Refer to the documentation of the `Callbacks` module for further information.
```
julia> import DifferentialRiccatiEquations.Callbacks
help?> Callbacks
```
Note that there are currently no pre-built observers.
## ADI shift parameter selection
The ADI shifts may be configured using keyword arguments of `solve`.
```julia
shifts = Shifts.Projection(2)
solve(::GALEProblem, ::ADI; shifts)
adi_kwargs = (; shifts)
solve(::GDREProblem, ::Ros1; adi_kwargs)
solve(::GAREProblem, ::NewtonADI; adi_kwargs)
```
Pre-built shift strategies include:
* `Heuristic` shifts described by [Penzl1999]
* `Projection` shifts described by [BennerKuerschnerSaak2014]
* User-supplied shifts via the `Cyclic` wrapper
Refer to the documentation of the `Shifts` module for further information.
```
julia> import DifferentialRiccatiEquations.Shifts
help?> Shifts
```
# Acknowledgments
I would like to thank the code reviewers:
* Jens Saak (https://github.com/drittelhacker)
* Martin Köhler (https://github.com/grisuthedragon)
* Fan Wang (https://github.com/FanWang00)
# License
The DifferentialRiccatiEquations package is licensed under [MIT], see `LICENSE`.
The `test/Rail371.mat` data file stems from [BennerSaak2005] and is licensed under [CC-BY-4.0].
See [MOR Wiki] for further information.
> **Warning**
> The output matrix `C` of the included configuration differs from all the other configurations hosted at [MOR Wiki] by a factor of 10.
[Penzl1999]: https://doi.org/10.1137/S1064827598347666
[BennerSaak2005]: http://nbn-resolving.de/urn:nbn:de:swb:ch1-200601597
[BennerKuerschnerSaak2014]: https://www.emis.de/journals/ETNA/vol.43.2014-2015/pp142-162.dir/pp142-162.pdf
[CC-BY-4.0]: https://spdx.org/licenses/CC-BY-4.0.html
[MIT]: https://spdx.org/licenses/MIT.html
[MOR Wiki]: http://modelreduction.org/index.php/Steel_Profile
| DifferentialRiccatiEquations | https://github.com/mpimd-csc/DifferentialRiccatiEquations.jl.git |
|
[
"MIT"
] | 0.1.2 | 8fe7c5bea08d2ea2c74df7d4a331e5c9db45167f | code | 613 | using SoleDecisionTreeInterface
using Documenter
DocMeta.setdocmeta!(SoleDecisionTreeInterface, :DocTestSetup, :(using SoleDecisionTreeInterface); recursive=true)
makedocs(;
modules=[SoleDecisionTreeInterface],
authors="Giovanni Pagliarini",
sitename="SoleDecisionTreeInterface.jl",
format=Documenter.HTML(;
canonical="https://aclai-lab.github.io/SoleDecisionTreeInterface.jl",
edit_link="main",
assets=String[],
),
pages=[
"Home" => "index.md",
],
)
deploydocs(;
repo="github.com/giopaglia/SoleDecisionTreeInterface.jl",
devbranch="main",
)
| SoleDecisionTreeInterface | https://github.com/aclai-lab/SoleDecisionTreeInterface.jl.git |
|
[
"MIT"
] | 0.1.2 | 8fe7c5bea08d2ea2c74df7d4a331e5c9db45167f | code | 3007 | module SoleDecisionTreeInterface
using Reexport
import DecisionTree as DT
@reexport using Sole
using Sole: DecisionTree
export solemodel
function solemodel(tree::DT.InfoNode, keep_condensed = false, use_featurenames = true, kwargs...)
# @show fieldnames(typeof(tree))
use_featurenames = use_featurenames ? tree.info.featurenames : false
root, info = begin
if keep_condensed
root = solemodel(tree.node; use_featurenames = use_featurenames, kwargs...)
info = (;
apply_preprocess=(y -> UInt32(findfirst(x -> x == y, tree.info.classlabels))),
apply_postprocess=(y -> tree.info.classlabels[y]),
)
root, info
else
root = solemodel(tree.node; replace_classlabels = tree.info.classlabels, use_featurenames = use_featurenames, kwargs...)
info = (;)
root, info
end
end
info = merge(info, (;
featurenames=tree.info.featurenames,
#
supporting_predictions=root.info[:supporting_predictions],
supporting_labels=root.info[:supporting_labels],
)
)
return DecisionTree(root, info)
end
# function solemodel(tree::DT.Root)
# root = solemodel(tree.node)
# # @show fieldnames(typeof(tree))
# info = (;
# n_feat = tree.n_feat,
# featim = tree.featim,
# supporting_predictions = root.info[:supporting_predictions],
# supporting_labels = root.info[:supporting_labels],
# )
# return DecisionTree(root, info)
# end
function solemodel(tree::DT.Node; replace_classlabels = nothing, use_featurenames = false)
test_operator = (<)
# @show fieldnames(typeof(tree))
feature = (use_featurenames != false) ? Sole.VariableValue(use_featurenames[tree.featid]) : Sole.VariableValue(tree.featid)
cond = ScalarCondition(feature, test_operator, tree.featval)
antecedent = Atom(cond)
lefttree = solemodel(tree.left; replace_classlabels = replace_classlabels, use_featurenames = use_featurenames)
righttree = solemodel(tree.right; replace_classlabels = replace_classlabels, use_featurenames = use_featurenames)
info = (;
supporting_predictions = [lefttree.info[:supporting_predictions]..., righttree.info[:supporting_predictions]...],
supporting_labels = [lefttree.info[:supporting_labels]..., righttree.info[:supporting_labels]...],
)
return Branch(antecedent, lefttree, righttree, info)
end
function solemodel(tree::DT.Leaf; replace_classlabels = nothing, use_featurenames = false)
# @show fieldnames(typeof(tree))
prediction = tree.majority
labels = tree.values
if !isnothing(replace_classlabels)
prediction = replace_classlabels[prediction]
labels = replace_classlabels[labels]
end
info = (;
supporting_predictions = fill(prediction, length(labels)),
supporting_labels = labels,
)
return SoleModels.ConstantModel(prediction, info)
end
end
| SoleDecisionTreeInterface | https://github.com/aclai-lab/SoleDecisionTreeInterface.jl.git |
|
[
"MIT"
] | 0.1.2 | 8fe7c5bea08d2ea2c74df7d4a331e5c9db45167f | code | 2295 | using Test
using MLJ
using MLJBase
using DataFrames
using MLJDecisionTreeInterface
using SoleDecisionTreeInterface
using Sole
X, y = @load_iris
X = DataFrame(X)
train_ratio = 0.8
train, test = partition(eachindex(y), train_ratio, shuffle=true)
X_train, y_train = X[train, :], y[train]
X_test, y_test = X[test, :], y[test]
println("Training set size: ", size(X_train), " - ", size(y_train))
println("Test set size: ", size(X_test), " - ", size(y_test))
println("Training set type: ", typeof(X_train), " - ", typeof(y_train))
println("Test set type: ", typeof(X_test), " - ", typeof(y_test))
Tree = MLJ.@load DecisionTreeClassifier pkg=DecisionTree
model = Tree(
max_depth=-1,
min_samples_leaf=1,
min_samples_split=2,
)
# Bind the model and data into a machine
mach = machine(model, X_train, y_train)
# Fit the model
fit!(mach)
sole_dt = solemodel(fitted_params(mach).tree)
@test SoleData.scalarlogiset(X_test; allow_propositional = true) isa PropositionalLogiset
# Make test instances flow into the model
apply!(sole_dt, X_test, y_test)
# apply!(sole_dt, X_test, y_test, mode = :append)
sole_dt = @test_nowarn @btime solemodel(fitted_params(mach).tree, true)
sole_dt = @test_nowarn @btime solemodel(fitted_params(mach).tree, false)
printmodel(sole_dt; max_depth = 7, show_intermediate_finals = true, show_metrics = true)
printmodel.(listrules(sole_dt, min_lift = 1.0, min_ninstances = 0); show_metrics = true);
printmodel.(listrules(sole_dt, min_lift = 1.0, min_ninstances = 0); show_metrics = true, show_subtree_metrics = true);
printmodel.(listrules(sole_dt, min_lift = 1.0, min_ninstances = 0); show_metrics = true, show_subtree_metrics= true, tree_mode=true);
readmetrics.(listrules(sole_dt; min_lift=1.0, min_ninstances = 0))
printmodel.(listrules(sole_dt, min_lift = 1.0, min_ninstances = 0); show_metrics = true);
interesting_rules = listrules(sole_dt; min_lift=1.0, min_ninstances = 0, custom_thresholding_callback = (ms)->ms.coverage*ms.ninstances >= 4)
# printmodel.(sort(interesting_rules, by = readmetrics); show_metrics = (; round_digits = nothing, ));
printmodel.(sort(interesting_rules, by = readmetrics); show_metrics = (; round_digits = nothing, additional_metrics = (; length = r->natoms(antecedent(r)))));
@test_broken joinrules(interesting_rules)
| SoleDecisionTreeInterface | https://github.com/aclai-lab/SoleDecisionTreeInterface.jl.git |
|
[
"MIT"
] | 0.1.2 | 8fe7c5bea08d2ea2c74df7d4a331e5c9db45167f | code | 566 | using SoleDecisionTreeInterface
using Test
using Random
function run_tests(list)
println("\n" * ("#"^50))
for test in list
println("TEST: $test")
include(test)
end
end
println("Julia version: ", VERSION)
test_suites = [
("Core", ["core.jl",]),
]
@testset "SoleDecisionTreeInterface.jl" begin
for ts in eachindex(test_suites)
name = test_suites[ts][1]
list = test_suites[ts][2]
let
@testset "$name" begin
run_tests(list)
end
end
end
println()
end
| SoleDecisionTreeInterface | https://github.com/aclai-lab/SoleDecisionTreeInterface.jl.git |
|
[
"MIT"
] | 0.1.2 | 8fe7c5bea08d2ea2c74df7d4a331e5c9db45167f | docs | 7653 | # SoleDecisionTreeInterface.jl
[](https://aclai-lab.github.io/SoleDecisionTreeInterface.jl/stable/)
[](https://aclai-lab.github.io/SoleDecisionTreeInterface.jl/dev/)
[](https://github.com/aclai-lab/SoleDecisionTreeInterface.jl/actions/workflows/CI.yml?query=branch%3Amain)
[](https://codecov.io/gh/giopaglia/SoleDecisionTreeInterface.jl)
<!--
[](https://cirrus-ci.com/github/giopaglia/SoleDecisionTreeInterface.jl)
-->
Ever wondered what to do with a trained decision tree? Start by inspecting its knowledge, and end up evaluating it in a dedicated framework!
This package allows you to convert learned [DecisionTree](https://github.com/JuliaAI/DecisionTree.jl) models to [Sole](https://github.com/aclai-lab/Sole.jl) decision tree models.
With a Sole model in your hand, you can then to treat the extracted knowledge in symbolic form, that is, as a set of logical formulas, which allows you to:
- Evaluate them in terms of
+ accuracy (e.g., confidence, lift),
+ relevance (e.g., support),
+ interpretability (e.g., syntax height, number of atoms);
- Modify them;
- Merge them.
<!-- Note: this is a newly developed package; its potential is still unknown. -->
## Usage
### Converting to a Sole model
```julia
using MLJ
using MLJDecisionTreeInterface
using DataFrames
X, y = @load_iris
X = DataFrame(X)
train, test = partition(eachindex(y), 0.8, shuffle=true);
X_train, y_train = X[train, :], y[train];
X_test, y_test = X[test, :], y[test];
# Train a model
learned_dt_tree = begin
Tree = MLJ.@load DecisionTreeClassifier pkg=DecisionTree
model = Tree(max_depth=-1, )
mach = machine(model, X_train, y_train)
fit!(mach)
fitted_params(mach).tree
end
using SoleDecisionTreeInterface
# Convert to Sole model
sole_dt = solemodel(learned_dt_tree)
```
### Model inspection & rule study
```julia-repl
julia> using Sole;
julia> # Make test instances flow into the model, so that test metrics can, then, be computed.
apply!(sole_dt, X_test, y_test);
julia> # Print Sole model
printmodel(sole_dt; show_metrics = true);
▣ V4 < 0.8
├✔ setosa : (ninstances = 7, ncovered = 7, confidence = 1.0, lift = 1.0)
└✘ V3 < 4.95
├✔ V4 < 1.65
│├✔ versicolor : (ninstances = 10, ncovered = 10, confidence = 1.0, lift = 1.0)
│└✘ V2 < 3.1
│ ├✔ virginica : (ninstances = 2, ncovered = 2, confidence = 1.0, lift = 1.0)
│ └✘ versicolor : (ninstances = 0, ncovered = 0, confidence = NaN, lift = NaN)
└✘ V3 < 5.05
├✔ V1 < 6.5
│├✔ virginica : (ninstances = 0, ncovered = 0, confidence = NaN, lift = NaN)
│└✘ versicolor : (ninstances = 0, ncovered = 0, confidence = NaN, lift = NaN)
└✘ virginica : (ninstances = 11, ncovered = 11, confidence = 0.91, lift = 1.0)
julia> # Extract rules that are at least as good as a random baseline model
interesting_rules = listrules(sole_dt, min_lift = 1.0, min_ninstances = 0);
julia> printmodel.(interesting_rules; show_metrics = true);
▣ (V4 < 0.8) ∧ (⊤) ↣ setosa : (ninstances = 30, ncovered = 7, coverage = 0.23, confidence = 1.0, natoms = 1, lift = 4.29)
▣ (¬(V4 < 0.8)) ∧ (V3 < 4.95) ∧ (V4 < 1.65) ∧ (⊤) ↣ versicolor : (ninstances = 30, ncovered = 10, coverage = 0.33, confidence = 1.0, natoms = 3, lift = 2.73)
▣ (¬(V4 < 0.8)) ∧ (V3 < 4.95) ∧ (¬(V4 < 1.65)) ∧ (V2 < 3.1) ∧ (⊤) ↣ virginica : (ninstances = 30, ncovered = 2, coverage = 0.07, confidence = 1.0, natoms = 4, lift = 2.5)
▣ (¬(V4 < 0.8)) ∧ (¬(V3 < 4.95)) ∧ (¬(V3 < 5.05)) ∧ (⊤) ↣ virginica : (ninstances = 30, ncovered = 11, coverage = 0.37, confidence = 0.91, natoms = 3, lift = 2.27)
julia> # Simplify rules while extracting and prettify result
interesting_rules = listrules(sole_dt, min_lift = 1.0, min_ninstances = 0, normalize = true);
julia> printmodel.(interesting_rules; show_metrics = true, syntaxstring_kwargs = (; threshold_digits = 2));
▣ V4 < 0.8 ↣ setosa : (ninstances = 30, ncovered = 7, coverage = 0.23, confidence = 1.0, natoms = 1, lift = 4.29)
▣ (V4 ∈ [0.8,1.65)) ∧ (V3 < 4.95) ↣ versicolor : (ninstances = 30, ncovered = 10, coverage = 0.33, confidence = 1.0, natoms = 2, lift = 2.73)
▣ (V4 ≥ 1.65) ∧ (V3 < 4.95) ∧ (V2 < 3.1) ↣ virginica : (ninstances = 30, ncovered = 2, coverage = 0.07, confidence = 1.0, natoms = 3, lift = 2.5)
▣ (V4 ≥ 0.8) ∧ (V3 ≥ 5.05) ↣ virginica : (ninstances = 30, ncovered = 11, coverage = 0.37, confidence = 0.91, natoms = 2, lift = 2.27)
julia> # Directly access rule metrics
readmetrics.(listrules(sole_dt; min_lift=1.0, min_ninstances = 0))
4-element Vector{NamedTuple{(:ninstances, :ncovered, :coverage, :confidence, :natoms, :lift), Tuple{Int64, Int64, Float64, Float64, Int64, Float64}}}:
(ninstances = 30, ncovered = 7, coverage = 0.23333333333333334, confidence = 1.0, natoms = 1, lift = 4.285714285714286)
(ninstances = 30, ncovered = 10, coverage = 0.3333333333333333, confidence = 1.0, natoms = 3, lift = 2.7272727272727275)
(ninstances = 30, ncovered = 2, coverage = 0.06666666666666667, confidence = 1.0, natoms = 4, lift = 2.5)
(ninstances = 30, ncovered = 11, coverage = 0.36666666666666664, confidence = 0.9090909090909091, natoms = 3, lift = 2.2727272727272725)
julia> # Show rules with an additional metric (syntax height of the rule's antecedent)
printmodel.(sort(interesting_rules, by = readmetrics); show_metrics = (; round_digits = nothing, additional_metrics = (; height = r->SoleLogics.height(antecedent(r)))));
▣ (V4 ≥ 1.65) ∧ (V3 < 4.95) ∧ (V2 < 3.1) ↣ virginica : (ninstances = 30, ncovered = 2, coverage = 0.06666666666666667, confidence = 1.0, height = 2, lift = 2.5)
▣ V4 < 0.8 ↣ setosa : (ninstances = 30, ncovered = 7, coverage = 0.23333333333333334, confidence = 1.0, height = 0, lift = 4.285714285714286)
▣ (V4 ∈ [0.8,1.65)) ∧ (V3 < 4.95) ↣ versicolor : (ninstances = 30, ncovered = 10, coverage = 0.3333333333333333, confidence = 1.0, height = 1, lift = 2.7272727272727275)
▣ (V4 ≥ 0.8) ∧ (V3 ≥ 5.05) ↣ virginica : (ninstances = 30, ncovered = 11, coverage = 0.36666666666666664, confidence = 0.9090909090909091, height = 1, lift = 2.2727272727272725)
julia> # Pretty table of rules and their metrics
metricstable(interesting_rules; metrics_kwargs = (; round_digits = nothing, additional_metrics = (; height = r->SoleLogics.height(antecedent(r)))))
┌────────────────────────────────────────┬────────────┬────────────┬──────────┬───────────┬────────────┬────────┬─────────┐
│ Antecedent │ Consequent │ ninstances │ ncovered │ coverage │ confidence │ height │ lift │
├────────────────────────────────────────┼────────────┼────────────┼──────────┼───────────┼────────────┼────────┼─────────┤
│ V4 < 0.8 │ setosa │ 30 │ 7 │ 0.233333 │ 1.0 │ 0 │ 4.28571 │
│ (V4 ∈ [0.8,1.65)) ∧ (V3 < 4.95) │ versicolor │ 30 │ 10 │ 0.333333 │ 1.0 │ 1 │ 2.72727 │
│ (V4 ≥ 1.65) ∧ (V3 < 4.95) ∧ (V2 < 3.1) │ virginica │ 30 │ 2 │ 0.0666667 │ 1.0 │ 2 │ 2.5 │
│ (V4 ≥ 0.8) ∧ (V3 ≥ 5.05) │ virginica │ 30 │ 11 │ 0.366667 │ 0.909091 │ 1 │ 2.27273 │
└────────────────────────────────────────┴────────────┴────────────┴──────────┴───────────┴────────────┴────────┴─────────┘
```
| SoleDecisionTreeInterface | https://github.com/aclai-lab/SoleDecisionTreeInterface.jl.git |
|
[
"MIT"
] | 0.1.2 | 8fe7c5bea08d2ea2c74df7d4a331e5c9db45167f | docs | 262 | ```@meta
CurrentModule = SoleDecisionTreeInterface
```
# SoleDecisionTreeInterface
Documentation for [SoleDecisionTreeInterface](https://github.com/giopaglia/SoleDecisionTreeInterface.jl).
```@index
```
```@autodocs
Modules = [SoleDecisionTreeInterface]
```
| SoleDecisionTreeInterface | https://github.com/aclai-lab/SoleDecisionTreeInterface.jl.git |
|
[
"MIT"
] | 0.1.0 | f836f161637142210db3aaa67300acb6e4af09fa | code | 3168 | module CodeTransformation
import Core: SimpleVector, svec, CodeInfo
import Base: uncompressed_ast, unwrap_unionall
export addmethod!, codetransform!
# Most of this code is derived from Nathan Daly's DeepcopyModules.jl
# which is under the MIT license.
# https://github.com/NHDaly/DeepcopyModules.jl
"""
jl_method_def(argdata, ci, mod) - C function wrapper
This is a wrapper of the C function with the same name, found in the Julia
source tree at julia/src/method.c
Use `addmethod!` or `codetransform!` instead of calling this function directly.
"""
jl_method_def(argdata::SimpleVector, ci::CodeInfo, mod::Module) =
ccall(:jl_method_def, Cvoid, (SimpleVector, Any, Ptr{Module}), argdata, ci, pointer_from_objref(mod))
# `argdata` is `svec(svec(types...), svec(typevars...))`
"Recursively get the typevars from a `UnionAll` type"
typevars(T::UnionAll) = (T.var, typevars(T.body)...)
typevars(T::DataType) = ()
@nospecialize # the below functions need not specialize on arguments
"Get the module of a function"
getmodule(F::Type{<:Function}) = F.name.mt.module
getmodule(f::Function) = getmodule(typeof(f))
"Create a call singature"
makesig(f::Function, args) = Tuple{typeof(f), args...}
"""
argdata(sig[, f])
Turn a call signature into the 'argdata' `svec` that `jl_method_def` uses
When a function is given in the second argument, it replaces the one in the
call signature.
"""
argdata(sig) = svec(unwrap_unionall(sig).parameters::SimpleVector, svec(typevars(sig)...))
argdata(sig, f::Function) = svec(svec(typeof(f), unwrap_unionall(sig).parameters[2:end]...), svec(typevars(sig)...))
"""
addmethod!(f, argtypes, ci)
Add a method to a function.
The types of the arguments is given as a `Tuple`.
Example:
```
g(x) = x + 13
ci = code_lowered(g)[1]
function f end
addmethod!(f, (Any,), ci)
f(1) # returns 14
```
"""
addmethod!(f::Function, argtypes::Tuple, ci::CodeInfo) = addmethod!(makesig(f, argtypes), ci)
"""
addmethod(sig, ci)
Alternative syntax where the call signature is a `Tuple` type.
Example:
```
addmethod!(Tuple{typeof(f), Any}, ci)
```
"""
function addmethod!(sig::Type{<:Tuple{F, Vararg}}, ci::CodeInfo) where {F<:Function}
jl_method_def(argdata(sig), ci, getmodule(F))
end
@specialize # restore default
"""
codetransform!(tr, dst, src)
Apply a code transformation function `tr` on the methods of a function `src`,
adding the transformed methods to another function `dst`.
Example: Search-and-replace a constant in a function.
```
g(x) = x + 13
function e end
codetransform!(g => e) do ci
for ex in ci.code
if ex isa Expr
map!(x -> x === 13 ? 7 : x, ex.args, ex.args)
end
end
ci
end
e(1) # returns 8
```
"""
function codetransform!(tr::Function, @nospecialize(dst::Function), @nospecialize(src::Function))
mod = getmodule(dst)
for m in methods(src).ms
ci = uncompressed_ast(m)
ci = tr(ci)
jl_method_def(argdata(m.sig, dst), ci, mod)
end
end
"Alternative syntax: codetransform!(tr, src => dst)"
codetransform!(tr::Function, @nospecialize(p::Pair{<:Function, <:Function})) =
codetransform!(tr, p.second, p.first)
end # module
| CodeTransformation | https://github.com/perrutquist/CodeTransformation.jl.git |
|
[
"MIT"
] | 0.1.0 | f836f161637142210db3aaa67300acb6e4af09fa | code | 1274 | using CodeTransformation
using Test
@testset "CodeTransformation.jl" begin
@test CodeTransformation.getmodule(typeof(sin)) === Base
@test CodeTransformation.getmodule(sin) === Base
let
# Test example from doctring to addmethod!
g(x) = x + 13
ci = code_lowered(g)[1]
function f end
addmethod!(f, (Any,), ci)
@test f(1) === 14
# Alternative syntax
function f2 end
@test CodeTransformation.makesig(f2, (Any,)) === Tuple{typeof(f2), Any}
addmethod!(Tuple{typeof(f2), Any}, ci)
@test f2(1) === 14
end
let
# Test example from doctring to codetransform!
g(x) = x + 13
function e end
codetransform!(g => e) do ci
for ex in ci.code
if ex isa Expr
map!(x -> x === 13 ? 7 : x, ex.args, ex.args)
end
end
ci
end
@test e(1) === 8
@test g(1) === 14
end
let
a = Vector{T} where T
b = CodeTransformation.typevars(a)
@test b isa Tuple
@test length(b) == 1
@test b[1] isa TypeVar
@test b[1].name == :T
@test b[1].lb === Union{}
@test b[1].ub === Any
end
end
| CodeTransformation | https://github.com/perrutquist/CodeTransformation.jl.git |
|
[
"MIT"
] | 0.1.0 | f836f161637142210db3aaa67300acb6e4af09fa | docs | 1714 | # CodeTransformation
[](https://travis-ci.com/perrutquist/CodeTransformation.jl)
[](https://ci.appveyor.com/project/perrutquist/CodeTransformation-jl)
[](https://codecov.io/gh/perrutquist/CodeTransformation.jl)
This is an experimental package for working with the [`CodeInfo`](https://pkg.julialang.org/docs/julia/THl1k/1.1.1/devdocs/ast.html#CodeInfo-1)
objects that are containded in the vectors that Julia's `code_lowered` and `code_typed` functions return.
These objects can be modified and then turned back into functions (technically methods),
making it possible to apply code transformations to functions defined in other packages,
or in Julia itself.
## Examples
Copy a method from one function to another via a `CodeInfo` object.
```julia
using CodeTransformation
g(x) = x + 13
ci = code_lowered(g)[1] # get the CodeInfo from g's first (and only) method
function f end # create an empty function that we can add a method to
addmethod!(Tuple{typeof(f), Any}, ci)
f(1) # returns 14
```
Search-and-replace in the function `g` from the previous example. (Applies to all
methods, but `g` only has one.)
```julia
function e end
codetransform!(g => e) do ci
for ex in ci.code
if ex isa Expr
map!(x -> x === 13 ? 7 : x, ex.args, ex.args)
end
end
ci
end
e(1) # returns 8
g(1) # still returns 14
```
Note: The syntax may change in the next minor release of this package.
| CodeTransformation | https://github.com/perrutquist/CodeTransformation.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 15273 | module Concepts
import LinearAlgebra
import Distributions
using Printf
#==============================================================================#
# Customized Exceptions #
#==============================================================================#
struct UnrecognizedSymbolException <: Exception
symbol_name::String
end
Base.showerror(io::IO,e::UnrecognizedSymbolException) =
@printf(io,"[%s]: Unrecognized symbol",e.symbol_name)
struct NotOverLoadedException <: Exception
fcn_name::String
end
Base.showerror(io::IO,e::NotOverLoadedException) =
@printf(io,"[%s]: Abstract method called. Concrete implementation is required.",e.fcn_name)
struct Unimplemented <: Exception
fcn_name::String
end
Base.showerror(io::IO,e::Unimplemented) =
@printf(io,"[%s]: Notimplemented",e.fcn_name)
export UnrecognizedSymbolException,
NotOverLoadedException,
Unimplemented
#==============================================================================#
# Customized Macros #
#==============================================================================#
# Serves as a annotator. So far it is sufficient. We can always add actual defi-
# nitions later if we want to enforce the property. I don't think Julia currently
# supports private functions.. It maybe possible with some dark-art style trick..
macro private() end
macro overload() end
macro abstract_instance() end
export @overload,
@abstract_instance,
@private
#==============================================================================#
# Type Declearations #
#==============================================================================#
const Optional{T} = Union{T,Nothing}
const VecOrMatOfNumbers = VecOrMat{<:Number}
const VecOrMatOfReals = VecOrMat{<:Real}
const VecOrMatOfFloats = VecOrMat{<:AbstractFloat}
const VecOrMatOfIntegers = VecOrMat{<:Integer}
const VecOrMatOf{T} = VecOrMat{<:T} where T<:Any
const MaybeMissing{T} = Union{Missing,T} where T<:Number
const AutoboxedArray{T} = Union{S,Array{S}} where S<:T
export Optional,
VecOrMatOfReals,
VecOrMatOfFloats,
VecOrMatOfIntegers,
VecOrMatOf,
MaybeMissing,
AutoboxedArray
global SYMBOL_LIST = Set([:Poisson,:Gaussian,:Gamma,:Bernoulli,:NegativeBinomial,
:Count,:Binary,
:dimension,:rank,:l2diff,:l2difference])
global AVAILABLE_DATA_LAYOUT = Set([:flatten,:bycol,:byrow,:asmatrix,:astensor])
#==============================================================================#
# Exponential Family #
#==============================================================================#
abstract type ExponentialFamily end
struct AbstractBernoulli <: ExponentialFamily end
struct AbstractBinomial <: ExponentialFamily end
struct AbstractGaussian <: ExponentialFamily end
struct AbstractPoisson <: ExponentialFamily end
struct AbstractGamma <: ExponentialFamily end
struct AbstractExponential <: ExponentialFamily end
struct AbstractNegativeBinomial <: ExponentialFamily end
struct AbstractGeometric <: ExponentialFamily end
export ExponentialFamily,
AbstractBernoulli,
AbstractBinomial,
AbstractGaussian,
AbstractGamma,
AbstractExponential,
AbstractNegativeBinomial,
AbstractPoisson,
forward_map
function forward_map() end
# function forward_map(distribution::T, args...;kwargs...) where T<:Any
# throw(NotOverLoadedException("forward_map"))
# end
#------------------------------------------------------------------------------#
struct Diagnostics{T<:Any} end
export Diagnostics
abstract type AbstractLoss end
export AbstractLoss
struct ErrorMetric end
struct LpSpace
p::Real
norm
function LpSpace(p::Real)
return new(p,
x -> LinearAlgebra.norm(x,p))
end
end
struct SchattenClass end
struct BoundedLinearOperator end
export LpSpace,
SchattenClass,
BoundedLinearOperator
#==============================================================================#
# Model Fitting #
#==============================================================================#
abstract type AbstractModelView end
abstract type AbstractFittingMethods end
#==============================================================================#
# Tracker #
#==============================================================================#
abstract type AbstractTracker end
abstract type AbstractView end
struct Continuous end
struct Categorical end
struct Binary end
struct Support end
export Continuous,
Categorical,
AbstractTracker,
Binary,
Support,
AbstractView
#==============================================================================#
# Sampling Models #
#==============================================================================#
abstract type AbstractSamplingModels end
export AbstractSamplingModels,
BernoulliModel,
UniformModel,
NonUniformModel
struct BernoulliModel <: AbstractSamplingModels
rate::AbstractFloat
function BernoulliModel()
# AbstractType Constructor
return new()
end
function BernoulliModel(rate::T) where T<:AbstractFloat
return new(rate);
end
end
struct UniformModel <: AbstractSamplingModels
rate::AbstractFloat
function UniformModel()
#Abstract Type Constructor
return new()
end
function UniformModel(rate::T) where T<:AbstractFloat
return new(rate)
end
end
struct NonUniformModel <: AbstractSamplingModels
rate::AbstractFloat
function NonUniformModel()
#Abstract Type Constructor
return new()
end
function NonUniformModel(rate::T) where T<:AbstractFloat
return new(rate)
end
end
#==============================================================================#
# Frequency Domain Objects #
#==============================================================================#
abstract type FrequencyDomainObjects end
abstract type AbstractMGF <: FrequencyDomainObjects end
export FrequencyDomainObjects,
AbstractMGF
#==============================================================================#
# Estimator #
#==============================================================================#
abstract type AbstractEstimator end
function estimator(of::T=nothing,arg...;kwargs...) where T<:Any
throw(NotOverLoadedException("estimator"))
end
export AbstractEstimator,
estimator
#==============================================================================#
# Comparator #
#==============================================================================#
abstract type AbstractComparator end
struct Comparator{T<:Any} <:AbstractComparator
field::Optional{Dict}
@abstract_instance
function Comparator{T}() where T<:Any
return new{T}(nothing)
end
function Comparator{T}(by::Type{T}) where T<:Any
return new{T}(nothing)
end
function Comparator{T}(by::T) where T<:Any
return new{T}(nothing)
end
function Comparator{T}(by::T;eval_at::Optional{AutoboxedArray{Real}}=nothing) where T<:AbstractMGF
return new{T}(Dict(:eval_at => eval_at))
end
end
const Comparator(of::T;eval_at::Optional{AutoboxedArray{Real}}=nothing) where T<:AbstractMGF = begin
Comparator{T}(of,eval_at=eval_at)
end
const Comparator(of::T) where T<:Any = begin
if !isa(of,Symbol)
if isa(of,Type)
Comparator{of}(of)
else
Comparator{T}(of)
end
else
Comparator(type_conversion(of))
end
end
export AbstractComparator,
Comparator
#==============================================================================#
# Random Structures #
#==============================================================================#
const UnivariateDistributions =
Distributions.Distribution{Distributions.Univariate,S} where S<:Distributions.ValueSupport
abstract type AbstractFixedRankMatrix end
export UnivariateDistributions,
AbstractFixedRankMatrix
#==============================================================================#
# Abstract Inteface #
#==============================================================================#
export provide,
predict,
check,
evaluate,
choose,
join,
disjoint_join,
groupby,
complete,
type_conversion
# convert,
function type_conversion() end
function complete() end
function groupby() end
function join() end
function disjoint_join() end
function provide(object::T=nothing,arg...;kwargs...) where T<:Any
throw(DomainError("[provide]: calling abstract method. Concrete implementation needed"))
return 0;
end
function is(is_object::T,arg...;kwargs...) where T<:Any
throw(DomainError("[is]: calling abstract method. Concrete implementation needed"))
end
function check()
throw(DomainError("[check]: calling abstract method. Concrete implementation needed"))
end
function pretty_print() end
# const Concepts.check(object::Symbol,arg...;kwargs::Optional{Any}...) = Concepts.check(Val{object},arg...;kwargs...)
# This is necessary due to compiler bug??
# @overload
# const Concepts.check(object::Symbol,arg1) = Concepts.check(Val{object},arg1)
@overload
const Concepts.check(object::Symbol,arg...;kwargs::Optional{Any}...) = Concepts.check(Val{object},arg...;kwargs...)
# This is necessary due to compiler bug??
# @overload
# const Concepts.check(object::Symbol,arg1) = Concepts.check(Val{object},arg1)
# @overload
# const Concepts.check(object::Symbol,arg1,arg2) = Concepts.check(Val{object},arg1,arg2)
# @overload
# const Concepts.check(object::Symbol,arg1,arg2,arg3) = Concepts.check(Val{object},arg1,arg2,arg3)
function predict(object::T=nothing,arg...;kwargs...) where T<:Any
throw(NotOverLoadedException("predict"))
end
function fit() end
function evaluate(object::T=nothing,arg...;kwargs...) where T<:Any
throw(NotOverLoadedException("evaluate"))
end
function choose() where T<:Any
throw(NotOverLoadedException("evaluate"))
end
@overload
const Concepts.choose(a::Symbol,b::Symbol;kwargs...) = Concepts.choose(Val{a},Val{b};kwargs...)
#==============================================================================#
# Base Overrides #
#==============================================================================#
# @overload
# function Base.convert(::Type{Float64}, x::Array{Float64, 1})
# return x[1]
# end
# @overload
# function Base.convert(::Type{Any}, x::Array{Float64, 1})
# return x[1]
# end
# @overload
# function Base.convert(::Type{Int64}, x::Array{Int64, 1})
# return x[1]
# end
# @overload
# function Base.convert(::Type{Any}, x::Array{Int64, 1})
# return x[1]
# end
# @overload
# function Base.convert(::Type{Any}, x::Array{T, 1}) where T<:Number
# if isempty(x)
# return T(0)
# else
# return x[1]
# end
# end
# @overload
# function Base.convert(::Type{MaybeMissing{S}}, x::VecOrMatOf{T}) where {T<:Any, S<:Any}
# ret = nothing;
# if isa(x, Vector)
# ret = Vector{MaybeMissing{S}}(undef,length(x))
# for i in 1:length(x)
# ret[i] = x[i]
# end
# end
# if isa(x,Matrix)
# row, col = size(x)
# ret = Matrix{MaybeMissing{S}}(undef,row,col)
# for i in row
# for j in col
# ret[i, j] = x[i, j]
# end
# end
# end
# return ret;
# end
# emacs indentation BUGGGGGGGGGG
# @overload
# function Base.convert(::Type{ExponentialFamily}, x::Symbol)
# if x == :Poisson || x == :Count
# return AbstractPoisson()
# end
# if x == :Gaussian || x == :Normal
# return AbstractGaussian()
# end
# if x == :Bernoulli || x == :Binary
# return AbstractBernoulli()
# end
# if x == :Gamma
# return AbstractGamma()
# end
# if x == :NegativeBinomial
# return AbstractNegativeBinomial()
# end
# throw(InexactError())
# end
@overload
function type_conversion(::Type{MaybeMissing{S}}, x::VecOrMatOf{T}) where {T<:Any, S<:Any}
ret = nothing;
if isa(x, Vector)
ret = Vector{MaybeMissing{S}}(undef,length(x))
for i in 1:length(x)
ret[i] = x[i]
end
end
if isa(x,Matrix)
row, col = size(x)
ret = Matrix{MaybeMissing{S}}(undef,row,col)
for i in row
for j in col
ret[i, j] = x[i, j]
end
end
end
return ret;
end
function type_conversion(::Type{Symbol}, x::Symbol)
return x
end
@overload
function type_conversion(::Type{ExponentialFamily}, x::Symbol)
if x == :Poisson || x == :Count
return AbstractPoisson()
end
if x == :Gaussian || x == :Normal
return AbstractGaussian()
end
if x == :Bernoulli || x == :Binary
return AbstractBernoulli()
end
if x == :Gamma
return AbstractGamma()
end
if x == :NegativeBinomial
return AbstractNegativeBinomial()
end
throw(InexactError())
end
@overload
function type_conversion(::Type{Symbol}, x::ExponentialFamily)
if x == AbstractPoisson()
return :Poisson
end
if x == AbstractGaussian()
return :Gaussian
end
if x == AbstractBernoulli()
return :Bernoulli
end
if x == AbstractGamma()
return :Gamma
end
if x == AbstractNegativeBinomial()
return :NegativeBinomial
end
end
@overload
function type_conversion(x::Symbol)
try return type_conversion(ExponentialFamily, x) catch end
try return type_conversion(FrequencyDomainObjects, x) catch end
throw(UnrecognizedSymbolException(String(x)))
end
# @overload
# function Base.convert(::Type{Symbol}, x::ExponentialFamily)
# if x == AbstractPoisson()
# return :Poisson
# end
# if x == AbstractGaussian()
# return :Gaussian
# end
# if x == AbstractBernoulli()
# return :Bernoulli
# end
# if x == AbstractGamma()
# return :Gamma
# end
# if x == AbstractNegativeBinomial()
# return :NegativeBinomial
# end
# end
# @overload
# function Base.convert(x::Symbol)
# try return Base.convert(ExponentialFamily,x) catch end
# try return Base.convert(FrequencyDomainObjects,x) catch end
# throw(UnrecognizedSymbolException(String(x)))
# end
end # Module: Concept
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 3005 | module Estimator
using ..Concepts
import Distributions
import StatsBase
export MLE,
ProfileLikelihood,
MOM
abstract type EstimationProcedure{T<:Any} end
## not super important
struct MLE{T<:Any} <:Concepts.AbstractEstimator
of::T
function MLE{T}() where T<:Any
return new{T}()
end
function MLE{T}(of::T) where T<:Any
return new{T}(of)
end
end
## not super important
struct MOM{T<:Any} <:Concepts.AbstractEstimator
of::T
function MOM{T}() where T<:Any
return new{T}()
end
function MOM{T}(of::T) where T<:Any
return new{T}(of)
end
end
struct ProfileLikelihood <: EstimationProcedure{MLE} end
function Concepts.estimator(name::MOM{AbstractNegativeBinomial}, data::AutoboxedArray{T}) where T<:Real
EX = StatsBase.mean(data)
VarX = StatsBase.var(data)
pₘₒₘ = EX / VarX
rₘₒₘ = EX * (pₘₒₘ) / (1 - pₘₒₘ)
return Dict{Symbol, Float64}(:p => pₘₒₘ, :r => rₘₒₘ)
end
# const MLE(of::Union{T,Symbol}) where T<:Any =
# !isa(of,Symbol) ? MLE{T}(of) : MLE(convert(of))
const MLE(of::Union{T,Symbol}) where T<:Any =
!isa(of,Symbol) ? MLE{T}(of) : MLE(type_conversion(of))
@overload
function Concepts.estimator(name::MLE{AbstractGaussian}, data::AutoboxedArray{T};
method::EstimationProcedure{MLE} = ProfileLikelihood()) where T<:Real
if method == ProfileLikelihood()
# for now, we use Distributions.jl. Will be replaced in the future.
est = Distributions.fit_mle(Distributions.Gaussian,data)
return Dict(:μ => est.μ, :σ => est.σ)
end
end
@overload
function Concepts.estimator(name::MLE{AbstractGamma},data::AutoboxedArray{T};
method::EstimationProcedure{MLE} = ProfileLikelihood()) where T<:Real
# default uses profile likelihood method
if method == ProfileLikelihood()
# for now, we use Distributions.jl. Will be replaced in the future.
est = Distributions.fit_mle(Distributions.Gamma,data)
return Dict(:α => est.α, :θ => est.θ)
end
end
# data layout needs to be implemented properly
@overload
function Concepts.estimator(name::MLE{AbstractPoisson},data::AutoboxedArray{T};
method::EstimationProcedure{MLE} = ProfileLikelihood(),
data_layout::Symbol = :flatten) where T<:Real
# default uses profile likelihood method
if method == ProfileLikelihood()
λ = sum(data)/length(data)
return Dict(:λ => λ)
end
end
@overload
function Concepts.estimator(name::MLE{AbstractBernoulli},data::AutoboxedArray{T};
method::EstimationProcedure{MLE} = ProfileLikelihood(),
data_layout::Symbol = :flatten) where T<:Real
# default uses profile likelihood method
if method == ProfileLikelihood()
est = Distributions.fit_mle(Distributions.Bernoulli,data)
return Dict(:p => est.p)
end
end
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 9889 | module Losses
#==============================================================================#
# MODULE OPTIONS & FLAGS #
const DEBUG_MODE = false
#==============================================================================#
const VERBOSE_MODE = true
#------------------------------------------------------------------------------#
using Printf
import Random, AutoGrad, Distributions
using ..Concepts
using ..Utilities.BatchUtils
# export BatchFactory
# struct SGD end
####################### HELPER METHODS ###################
# vectorized sigmoid function
σ(z) = 1.0 ./ (1.0 .+ exp.(-z))
##########################################################
# function Poisson()
# L(x,y,c,ρ) = sum(exp.(x) .- y .* x) + sum(ρ .* (x .- c).^2)
# return L;
# end
# function Logistic()
# L(x,y,c,ρ) = -sum(y .* log.(σ.(x)) .+ (1 .- y) .* log.(1 .- σ.(x))) .+ sum(ρ .* (x .- c).^2);
# return L;
# end
export Loss,
train,
sgd_train,
subgrad_train,
negative_binomial_train
struct Loss{T} <: AbstractLoss where T<:Any
function Loss{T}() where T<:Any
@abstract_instance
return new{T}()
end
# function Loss{AbstractPoisson}(of::Union{AbstractPoisson,Type{Val{:Poisson}}})
function Loss{T}(of::T) where T<:ExponentialFamily
return new{T}()
end
end
# kind of hackish.. the second one just defined in the first.
const Loss(of::Union{T,Symbol}) where T<:ExponentialFamily =
typeof(of) <: ExponentialFamily ? Loss{T}(of) : Loss(type_conversion(ExponentialFamily, of))
loss_logistic(x,y,c,ρ) = -sum(y .* log.(σ.(x)) .+ (1 .- y) .* log.(1 .- σ.(x))) .+ sum(ρ .* (x .- c).^2);
#==============================================================================#
# Gaussian Loss #
#==============================================================================#
function Concepts.provide(loss::Loss{AbstractGaussian})
#TODO
end
function Concepts.evaluate(loss::Loss{AbstractGaussian},
x,y,c,ρ)
#TODO
return sum(0.5 .* (y .- x).^2) + sum(ρ .* (x .- c).^2);
end
function grad(loss::Loss{AbstractGaussian} ,
x,y,c,ρ)
#TODO
return -(y .- x) .+ (2*ρ) .* (x .- c)
end
#==============================================================================#
# Bernoulli (Logistic) Loss #
#==============================================================================#
function Concepts.provide(loss::Loss{AbstractBernoulli})
L(x,y,c,ρ) = -sum(y .* log.(σ.(x)) .+ (1 .- y) .* log.(1 .- σ.(x))) .+ sum(ρ .* (x .- c).^2);
return L
end
function Concepts.evaluate(loss::Loss{AbstractBernoulli},
x,y,c,ρ)
return -sum(y .* log.(σ.(x)) .+ (1 .- y) .* log.(1 .- σ.(x)))
.+ sum(ρ .* (x .- c).^2);
end
function grad(loss::Loss{AbstractBernoulli},
x,y,c,ρ)
ex = exp.(x)
inv_ex1 = 1 ./(ex .+ 1)
return inv_ex1 .* (-y + (1 .-y) .* ex) .+ (2*ρ) .* (x .- c)
end
#==============================================================================#
# Poisson Loss #
#==============================================================================#
@overload
function Concepts.provide(loss::Loss{AbstractPoisson})
L(x,y,c,ρ) = sum(exp.(x) .- y .* x) + sum(ρ .* (x .- c).^2)
return L;
end
function Concepts.evaluate(loss::Loss{AbstractPoisson},
x,y,c,ρ)
return sum(exp.(x) .- y .* x) + sum(ρ .* (x .- c).^2)
end
function grad(loss::Loss{AbstractPoisson},
x,y,c,ρ)
return exp.(x) .- y .+ (2*ρ) .* (x .- c)
end
#==============================================================================#
# Gamma Loss #
#==============================================================================#
function Concepts.provide(loss::Loss{AbstractGamma})
L(x,y,c,ρ) = sum(-x .* y .- log.(-x))+ sum(ρ .* (x .- c).^2)
return L
end
# function Concepts.evaluate(loss::Loss{AbstractGamma},x,y,c,ρ)
# return sum(x .* y .- log.(x))+ sum(ρ .* (x .- c).^2)
# end
function Concepts.evaluate(loss::Loss{AbstractGamma},x,y,c,ρ)
return sum(y .* exp.(x) .- x) + sum(ρ .* (x .- c).^2)
end
# function _evaluate(loss::Loss{AbstractGamma},x,y,c,ρ)
# x₋ = x[findall(a -> a < 0, x)]
# y₋ = y[findall(a -> a < 0, x)]
# x₊ = x[findall(a -> a > 0, x)]
# y₊ = x[findall(a -> a > 0, x)]
# return (-1) * sum(x₋ .* y₋ .+ log.(-x₋)) + (-1) * sum(-x₊ .* y₊ .+ log.(x₊)) + sum(ρ .* (x .- c).^2)
# end
## Use the reciprocal link instead of the negative reciprocal link
# function grad(loss::Loss{AbstractGamma}, x, y, c, ρ)
# return y .- (1 ./ x) .+ (2*ρ) .* (x .- c)
# end
function grad(loss::Loss{AbstractGamma}, x, y, c, ρ)
return y .* exp.(x) .- 1 + (2*ρ) .* (x .- c)
end
function subgrad(loss::Loss{AbstractGamma}, x, y, c, ρ)
∇ = zeros(length(x))
pos_id = findall(a -> a > 0, x)
neg_id = findall(a -> a < 0, x)
∇[neg_id] = (-1) .* (y[neg_id] - (1 ./ x[neg_id])) .+ (2*ρ) .* (x[neg_id] .- c[neg_id])
∇[pos_id] = (-1) .* (-y[pos_id] + (1 ./ x[pos_id])) .+ (2*ρ) .* (x[pos_id] .- c[pos_id])
return ∇
end
function grad_logistic(x,y,c,ρ)
ex = exp.(x)
inv_ex1 = 1 ./(ex .+ 1);
return inv_ex1 .* (-y + (1 .-y) .* ex) .+ (2*ρ) .* (x .- c);
# return (-y .* inv_ex1 + (1 .- y) .* (ex .* inv_ex1)) .+ (2*ρ) .* (x.-c);
end
#==============================================================================#
# Negative Binomial Loss #
#==============================================================================#
function Concepts.provide(loss::Loss{AbstractNegativeBinomial})
## to implement
return nothing
end
function Concepts.evaluate(loss::Loss{AbstractNegativeBinomial}, x, y, c, ρ; r_estimate)
return sum(y .* exp.(x) - r_estimate .* log.(1 .- exp.(-exp.(x)))) .+ sum(ρ .* (x .- c).^2)
# originally it is. However, this is a constrained optimization problem.
# return sum(-y .* x .- r_estimate .* log.(1 .- exp.(x))) + sum(ρ .* (x .- c).^2)
end
function grad(loss::Loss{AbstractNegativeBinomial}, x, y, c, ρ; r_estimate = nothing)
return y .* exp.(x) .- r_estimate .* exp.(x) ./ (exp.(exp.(x)) .- 1) + (2*ρ) .* (x .- c)
end
function train(loss;fx,y,c,ρ,γ=0.02,iter=20,verbose=false)
DEBUG_MODE && @info "Gradient Descent with Autograd"
∇ = AutoGrad.grad(loss);
curFx = fx;
for i = 1:iter
curFx = curFx .- γ .* ∇(curFx,y,c,ρ);
end
return curFx;
end
function train(native_loss::Loss{T};
fx, y, c, ρ, γ=0.02, iter=20, verbose=false, subgrad = false) where T<:ExponentialFamily
DEBUG_MODE && @info "Gradient Descent with native differentitaion"
curFx = fx;
for i = 1:iter
curFx .-= γ * grad(native_loss, curFx, y, c, ρ);
# curFx .-= γ * grad(native_loss, curFx, y, c, ρ);
# if project == true
# curFx = abs.(curFx)
# end
if verbose == true
@printf("loss:%f\n", Concepts.evaluate(native_loss,curFx,y,c,ρ ))
end
end
return curFx;
end
# specialization for negative binomial loss
function negative_binomial_train(;fx, y, c, ρ, γ=0.02, iter=20, verbose=false, r_estimate = nothing)
DEBUG_MODE && @info "Gradient Descent with native differentitaion"
curFx = fx;
for i = 1:iter
# @show("here")
curFx .-= γ * grad(Loss{AbstractNegativeBinomial}(), curFx, y, c, ρ;r_estimate = r_estimate);
if verbose == true
@printf("loss:%f\n", Concepts.evaluate(Loss{AbstractNegativeBinomial}(),
curFx,y,c,ρ; r_estimate = r_estimate))
end
end
return curFx;
end
function subgrad_train(native_loss::Loss{T};
fx, y, c, ρ, γ=0.02, iter=20, verbose=false) where T<:ExponentialFamily
DEBUG_MODE && @info "Gradient Descent with native differentitaion"
curFx = fx;
for i = 1:iter
curFx .-= γ * subgrad(native_loss, curFx, y, c, ρ);
if verbose == true
@printf("loss:%f\n",_evaluate(native_loss,curFx,y,c,ρ ))
end
end
return curFx;
end
function sgd_train(native_loss::Loss{T};
fx, y, c, ρ, α, ρ₁, ρ₂, batch_size, epoch) where T<:ExponentialFamily
n = length(fx)
curFx = fx
batch = BatchFactory{SequentialScan}(size = batch_size)
initialize(batch, fx)
s = zeros(batch_size)
r = zeros(batch_size)
ŝ = zeros(batch_size)
r̂ = zeros(batch_size)
for i in 1:epoch
while has_next(batch)
cur_batch = next(batch)
∇ₛₐₘₚₗₑ = grad(native_loss, curFx[cur_batch], y[cur_batch], c[cur_batch], ρ)
s .= (ρ₁ .* s) .+ (1 .- ρ₁) .* ∇ₛₐₘₚₗₑ
r .= (ρ₂ .* r) .+ (1 .- ρ₂) .* (∇ₛₐₘₚₗₑ.^2)
ŝ .= s ./ (1 - ρ₁^i)
r̂ .= r ./ (1 - ρ₂^i)
# @show(r̂)
curFx[cur_batch] = curFx[cur_batch] - α ./ sqrt.(r̂) .* ŝ
end
# @show(Concepts.evaluate(native_loss,curFx,y,c,ρ))
reset(batch)
end
return curFx
end
function sgd_subgrad_train(native_loss::Loss{T};
fx, y, c, ρ, α, ρ₁, ρ₂, batch_size, epoch) where T<:ExponentialFamily
n = length(fx)
curFx = fx
batch = BatchFactory{SequentialScan}(size = batch_size)
initialize(batch, fx)
s = zeros(batch_size)
r = zeros(batch_size)
ŝ = zeros(batch_size)
r̂ = zeros(batch_size)
for i in 1:epoch
while has_next(batch)
cur_batch = next(batch)
∇ₛₐₘₚₗₑ = subgrad(native_loss, curFx[cur_batch], y[cur_batch], c[cur_batch], ρ)
s .= (ρ₁ .* s) .+ (1 .- ρ₁) .* ∇ₛₐₘₚₗₑ
r .= (ρ₂ .* r) .+ (1 .- ρ₂) .* (∇ₛₐₘₚₗₑ.^2)
ŝ .= s ./ (1 - ρ₁^i)
r̂ .= r ./ (1 - ρ₂^i)
# @show(r̂)
curFx[cur_batch] = curFx[cur_batch] - α ./ sqrt.(r̂) .* ŝ
end
# @show(Concepts.evaluate(native_loss,curFx,y,c,ρ))
reset(batch)
end
return curFx
end
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 5420 | module BetterMGF
using ..Concepts
import LinearAlgebra
struct MGF{T<:Any} <: AbstractMGF
OPTION_LOG_SCALE::Optional{Bool}
function MGF{T}() where T<:Any
return new{T}()
end
function MGF{T}(object::T;logscale::Optional{Bool} = nothing) where T<:Any
if isnothing(logscale)
return new{T}()
end
return new{T}(logscale)
end
end
const MGF() = MGF{Any}()
const MGF(object::Union{T, Symbol}; logscale::Optional{Bool} = nothing) where T<:Any =
!isa(object,Symbol) ? MGF{T}(object, logscale = logscale) : MGF(type_conversion(object), logscale=logscale)
struct SampleMGF <: AbstractMGF
OPTION_LOG_SCALE::Optional{Bool}
function SampleMGF(;logscale::Optional{Bool} = nothing)
if isnothing(logscale)
return new()
end
return new(logscale)
end
end
export MGF,
SampleMGF
# @overload
# function Base.convert(::Type{FrequencyDomainObjects},x::Symbol)
# if x == :MGF
# return MGF
# end
# end
@overload
function Concepts.type_conversion(::Type{FrequencyDomainObjects}, x::Symbol)
if x == :MGF
return MGF
end
end
@overload
function Concepts.evaluate(object::MGF{AbstractPoisson},t::AutoboxedArray{Real};
λ::S) where S<:Real
if λ <= 0
throw(DomainError("λ should be a positive number"))
end
if !isnothing(object.OPTION_LOG_SCALE) && object.OPTION_LOG_SCALE == true
@warn "[evaluate(MGF{Poisson}): using log-scale]"
return λ .* (exp.(t) .- 1)
end
return exp.(λ .* (exp.(t) .- 1))
end
@overload
function Concepts.evaluate(object::MGF{AbstractBernoulli},t::AutoboxedArray{Real};
p::S) where S<:Real
if p < 0 || p >1
throw(DomainError("p should be a positive number in [0,1]"))
end
if !isnothing(object.OPTION_LOG_SCALE) && object.OPTION_LOG_SCALE == true
@warn "[evaluate(MGF{Bernoulli}): using log-scale]"
return log.((1 .- p) .+ p .* exp.(t))
end
return (1 .- p) .+ p .* exp.(t)
end
@overload
function Concepts.evaluate(object::MGF{AbstractGamma},t::AutoboxedArray{Real};
α::S,θ::S) where S<:Real
if α <= 1e-5 || θ <= 1e-5
throw(DomainError("α and θ should be a positive number"))
end
if !isnothing(object.OPTION_LOG_SCALE) && object.OPTION_LOG_SCALE == true
@warn "[evaluate(MGF{Gamma}): using log-scale]"
return (-α) .* log.(1 .- t .* θ)
end
return (1 .- t .* θ) .^ (-α)
end
@overload
function Concepts.evaluate(object::MGF{AbstractGaussian},t::AutoboxedArray{Real};
μ::S = 0,σ::S=1) where S<:Real
if abs(σ) <= 1e-4
throw(DomainError("σ should be non-zero. Otherwise, it will be degenerate."))
end
if !isnothing(object.OPTION_LOG_SCALE) && object.OPTION_LOG_SCALE == true
@warn "[evaluate(MGF{Gaussian}): using log-scale]"
return t .* μ .+ σ^2 / 2 .* t.^2
end
return exp.(t .* μ .+ σ^2 / 2 .* t.^2)
end
import Distributions
@overload
function Concepts.evaluate(object::MGF{AbstractNegativeBinomial},t::AutoboxedArray{Real};
r::S = 0,p::S=1) where S<:Real
if r <= 1e-5 || p <= 1e-5 || p >=1
throw(DomainError("[Negative Binomial]: r should be > 0, p should be in (0,1)"))
end
if !isnothing(object.OPTION_LOG_SCALE) && object.OPTION_LOG_SCALE == true
@warn "[evaluate(MGF{NegativeBinomial})]: using log-scale"
return (r .* (log.(1 .- p) .+ t)) .- (r .* log.(1 .- p .* exp.(t)))
end
return ((1 .- p) .* exp.(t)).^r ./ (1 .- p .* exp.(t)).^r
end
@overload
function Concepts.evaluate(object::SampleMGF,t::VecOrMat{T};data::VecOrMatOf{Real},order::Integer = 8,data_layout::Symbol=:flatten) where T <: Real
if isa(t,Matrix) || isa(data,Matrix)
# throw(DomainError("Unimplemented"))
if data_layout == :flatten
@info "MGF of the matrix is evaluated by flattening the data matrix into a vector"
return dp_get_sample_basis(t,order) * dp_get_sample_mean(data[:],order)
end
if data_layout == :bycol
throw(DomainError("Unimplemented"))
end
if data_layout == :byrow
throw(DomainError("Unimplemented"))
end
end
return dp_get_sample_basis(t,order) * dp_get_sample_mean(data,order)
end
@private
function dp_get_denominator(order::Integer = 10)
denom = ones(order+1)
for i = 2:order+1
denom[i] = denom[i-1]*(i-1)
end
return 1 ./ denom
end
@private
function dp_get_numerator(t::Array{S,1},order::Integer = 10) where S<:Real
n = length(t)
T = ones(n,order+1)
for i = 2: order +1
T[:,i] = T[:,i-1] .* t
end
return T
end
@private
function dp_get_sample_basis(t::Array{S,1}, order::Integer = 10) where S<:Real
num = dp_get_numerator(t,order)
denom = dp_get_denominator(order)
for i = 1:size(num)[1]
num[i,:] = num[i,:] .* denom
end
return num
end
@private
function dp_get_sample_mean(data::Array{S,1}, order::Integer = 10) where S<: Real
ret = ones(order + 1)
copy_of_data = deepcopy(data)
n = length(copy_of_data)
for i = 2:order + 1
ret[i] = sum(copy_of_data)/n
copy_of_data = copy_of_data .* data
end
return ret
end
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 5533 | module MatrixCompletion
global VERBOSE_MODE = true
global DEBUG_MODE = true
macro api(as,of)
eval(Expr(:toplevel,:($as=$of),:(export $as)))
end
function dbg(x)
if VERBOSE_MODE == true
@show x
end
end
function see(x)
if VERBOSE_MODE == true
display(x)
end
end
function format(str)
return rpad(str,70,".")
end
export dbg,see,format
include("./Concepts.jl")
include("./MGF.jl")
include("./Estimator.jl")
include("./ModelFitting.jl")
include("./Utilities/Utilities.jl")
include("./Losses.jl")
include("./Library/MathLibSignatures.jl")
include("./Library/MathLib.jl")
@api MatrixCompletionModel ModelFitting.MatrixCompletionModel
@api BatchFactory Utilities.BatchUtils.BatchFactory
@api BatchingStrategy Utilities.BatchUtils.BatchingStrategy
@api SequentialScan Utilities.BatchUtils.SequentialScan
@api MaybeMissing Concepts.MaybeMissing
@api VecOrMatOf Concepts.VecOrMatOf
@api UnivariateDistributions Concepts.UnivariateDistributions
@api eigs Utilities.FastEigen.eigs
@api NativeLOBPCG Utilities.FastEigen.NativeLOBPCG
@api NativeEigen Utilities.FastEigen.NativeEigen
@api KrylovMethods Utilities.FastEigen.KrylovMethods
@api groupby Concepts.groupby
@api join Concepts.join
@api provide Concepts.provide
@api check Concepts.check
@api predict Concepts.predict
@api evaluate Concepts.evaluate
@api estimator Concepts.estimator
@api choose Concepts.choose
@api complete Concepts.complete
@api NotOverLoadedException Concepts.NotOverLoadedException
@api UnrecognizedSymbolException Concepts.UnrecognizedSymbolException
#==============================================================================#
# Model Fitting #
#==============================================================================#
@api AbstractModelView Concepts.AbstractModelView
#==============================================================================#
# Comparator #
#==============================================================================#
@api Comparator Concepts.Comparator
#==============================================================================#
# Loss Functions #
#==============================================================================#
@api AbstractLoss Concepts.AbstractLoss
@api Loss Losses.Loss
@api train Losses.train
#==============================================================================#
# Estimator #
#==============================================================================#
#@api EstimationProcedure Estimator.EstimationProcedure
@api ProfileLikelihood Estimator.ProfileLikelihood
@api MLE Estimator.MLE
@api MOM Estimator.MOM
#==============================================================================#
# Exponential Family #
#==============================================================================#
@api ExponentialFamily Concepts.ExponentialFamily
@api Gamma Concepts.AbstractGamma
@api Binomial Concepts.AbstractBinomial
@api Gaussian Concepts.AbstractGaussian
@api Poisson Concepts.AbstractPoisson
@api Bernoulli Concepts.AbstractBernoulli
@api NegativeBinomial Concepts.AbstractNegativeBinomial
@api GaussianMatrix Utilities.GaussianMatrix
@api PoissonMatrix Utilities.PoissonMatrix
@api BernoulliMatrix Utilities.BernoulliMatrix
@api GammaMatrix Utilities.GammaMatrix
@api forward_map Concepts.forward_map
@api AbstractSamplingModels Concepts.AbstractSamplingModels
@api AbstractFixedRankMatrix Concepts.AbstractFixedRankMatrix
@api FixedRankMatrix Utilities.FixedRankMatrix
#==============================================================================#
# Tracker #
#==============================================================================#
@api IndexTracker Utilities.IndexTracker
@api disjoint_join Concepts.disjoint_join
@api Continuous Concepts.Continuous
@api Categorical Concepts.Categorical
@api MGF BetterMGF.MGF
@api SampleMGF BetterMGF.SampleMGF
@api Sampler Utilities.Sampler
@api BernoulliModel Concepts.BernoulliModel
@api UniformModel Concepts.UniformModel
# @api NonUniformModel Concepts.NonUniformModel
@api Diagnostics Concepts.Diagnostics
@api LpSpace Concepts.LpSpace
@api ErrorMetric Concepts.ErrorMetric
@api RelativeError Utilities.RelativeError
@api AbsoluteError Utilities.AbsoluteError
@api within_radius Utilities.within_radius
# include("./NonConvex/lowrankmodels/LowRankModels.jl")
include("./Convex/ADMM.jl")
include("./NonConvex/chained_glrm.jl")
@api OneShotADMM ADMM.OneShotADMM
@api ChainedADMM ADMM.ChainedADMM
@api ChainedALM ALM.ChainedALM
@api OneShotALM ALM.OneShotALM
@api ProxGradParams LowRankModels.ProxGradParams
# @api complete ADMM.complete
end # module
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 9619 | module ModelFitting
using ..Concepts
using ..BetterMGF
using ..Estimator
import LinearAlgebra
export DistributionDeduction
export ObservedOrMissing
export MatrixCompletionModel
struct DistributionDeduction <: Concepts.AbstractModelView end
struct ObservedOrMissing <: Concepts.AbstractModelView end
struct MatrixCompletionModel end
@overload
function Concepts.check(object::Union{Type{Val{:continuous}},Continuous},
data::Array{T}) where T<:Real
dist_to_integral_cast = LinearAlgebra.norm(data .- round.(data),2)
return dist_to_integral_cast > 1e-2
end
@overload
function Concepts.check(object::Union{Type{Val{:integral}},Categorical},
data::Array{T}) where T<:Real
dist_to_integral_cast = LinearAlgebra.norm(data .- round.(data),2)
return dist_to_integral_cast <= 1e-4
end
@overload
function Concepts.check(object::Union{Type{Binary},
Type{Val{:binary}},
Binary},
data::Array{T}) where T<:Real
return length(unique(data)) == 2
end
# const Concepts.check(object::Symbol,data::Array{T}) where T<:Real = Concepts.check(Val{object},data)
# @overload
# const Concepts.check(object::Symbol,arg...;kwargs...) = Concepts.check(Val{object},arg...;kwargs...)
@overload
function Concepts.check(object::Union{Type{Support},
Type{Val{:support}},
Support},
data::Array{T};
layout::Symbol = :flatten) where T<:Real
if layout == :flatten
return (minimum(data[:]), maximum(data[:]))
end
# TODO: implement for other data layouts
end
#const Concepts.check(object::Symbol,data::Array{T;layout::Symbol =:flatten) where T<:Real =
# Concepts.check(Val{object},data,layout=layout)
# for now we only compare with Gamma. More smooth distributions will be added later.
@overload
function Concepts.check(object::Union{Type{Val{:Gaussian}},
Type{AbstractGaussian},
AbstractGaussian},
data::Array{T}) where T<:Real
if check(:continuous,data) == false
return false
end
# check against gamma when we get positive support
if check(:support,data) > (0,0) && choose(Gaussian,Gamma) == :Gamma
return false
end
return true
end
@overload
function Concepts.check(object::Union{Type{Val{:Gamma}},
Type{AbstractGamma},
AbstractGamma},
data::Array{T}) where T<:Real
if check(:continuous,data) == false
return false
end
if check(:support,data) < (0,0)
return false
end
if choose(Gaussian,Gamma) == :Gaussian
return false
end
return true
end
# For now we let non binary categorical array be Poisson. We will refine this later.
@overload
function Concepts.check(object::Union{Type{Val{:Poisson}},
Type{AbstractPoisson},
AbstractPoisson},
data::Array{T}) where T<:Real
if check(:integral,data) == false
return false
end
if check(:binary,data) == true
return false
end
return true
end
@overload
function Concepts.check(object::Union{Type{Val{:Bernoulli}},
Type{AbstractBernoulli},
AbstractBernoulli},
data::Array{T}) where T<:Real
if check(:integral,data) == false
return false
end
if check(:binary,data) == false
return false
end
return true
end
@overload
function Concepts.check(object::Union{Type{Val{:NegativeBinomial}},
Type{AbstractNegativeBinomial},
AbstractNegativeBinomial},
data::Array{T}) where T<:Real
return false #TODO: implement this properly
end
@overload
function Concepts.choose(a::Union{AbstractPoisson,
Type{AbstractPoisson},
Type{Val{:Poisson}}},
b::Union{AbstractNegativeBinomial,
Type{AbstractNegativeBinomial},
Type{Val{:NegativeBinomial}}};
data::AutoboxedArray{Real} = nothing,
comp::Comparator = Comparator{MGF}(MGF()))
# for now we return poisson all the time
return :Poisson
end
# we choose to return symbol for maximum flexibility
@overload
function Concepts.choose(a::Union{AbstractGaussian,
Type{AbstractGaussian},
Type{Val{:Gaussian}}},
b::Union{AbstractGamma,
Type{AbstractGamma},
Type{Val{:Gamma}}};
data::AutoboxedArray{Real} = nothing,
comp::Comparator = Comparator{MGF}(MGF()))
if check(:support,data) <= (0,)
@info "Data has negative support, force cast to Gaussian"
return :Gaussian
end
mle_est_gaussian = estimator(MLE{AbstractGaussian}(),data)
mle_est_gamma = estimator(MLE{AbstractGamma}(),data)
t = nothing
if isnothing(comp.field[:eval_at])
if mle_est_gamma[:α] <= 15 && mle_est_gamma[:θ] <= 15
t = collect(0.01:0.001:0.02)
else
t = collect(0:0.0001:0.001)
end
else
t = comp.field[:eval_at]
end
empirical_mgf = evaluate(SampleMGF(),t, data=data, order=20)
gaussian_mgf = evaluate(MGF(:Gaussian),t,μ = mle_est_gaussian[:μ],σ=mle_est_gaussian[:σ])
gamma_mgf = evaluate(MGF(:Gamma),t,α=mle_est_gamma[:α],θ=mle_est_gamma[:θ])
if check(:l2diff,empirical_mgf, gaussian_mgf) < check(:l2diff,empirical_mgf,gamma_mgf)
return :Gaussian
end
return :Gamma
end
@overload
function Concepts.fit(model::DistributionDeduction, data = Array{MaybeMissing{T}, 2};
data_layout::Symbol = :by_col) where T<: Number
distribution_view = Array{Symbol, 2}(undef, size(data))
if data_layout == :by_col
for i = 1:size(data)[2]
current_column = convert(Array{Float64}, filter(x -> !ismissing(x), data[:, i]))
# @show(typeof(current_column))
if check(:continuous, current_column)
distribution_view[:, i] .= choose(:Gaussian, :Gamma, data = current_column)
# display(distribution_view[:, i])
end
if check(:integral, current_column)
# @show(i)
if check(:binary, current_column)
distribution_view[:, i] .= :Bernoulli
end
if check(:Poisson, current_column)
distribution_view[:, i] .= :Poisson
end
end
end
end
if data_layout == :by_row
# TODO
end
# @show(distribution_view)
return distribution_view
end
@overload
function Concepts.fit(model::ObservedOrMissing, data = Array{MaybeMissing{T}, 2})
data_view = Array{Symbol, 2}(undef, size(data))
for r in 1:size(data)[1]
for c in 1:size(data)[2]
if ismissing(data[r, c])
data_view[r, c] = :Missing
else
data_view[r, c] = :Observed
end
end
end
return data_view
end
@overload
function Concepts.predict(model::MatrixCompletionModel;
completed_matrix,
type_tracker,
estimators = nothing)
predicted_matrix = similar(completed_matrix)
for dist in setdiff(keys(type_tracker.indices), [:Missing, :Observed])
# idx = type_tracker[convert(Symbol, dist)]
idx = type_tracker[type_conversion(Symbol, dist)]
# if convert(Symbol, dist) != :NegativeBinomial
if type_conversion(Symbol, dist) != :NegativeBinomial
predicted_matrix[idx] .= predict(dist, forward_map(Val{dist}, completed_matrix[idx]))
else
predicted_matrix[idx] .= predict(dist, forward_map(Val{dist}, completed_matrix[idx],
r_estimate = estimators[:NegativeBinomial][:r]))
end
end
return predicted_matrix
end
@overload
function Base.summary(model::MatrixCompletionModel;
predicted_matrix,
truth_matrix,
type_tracker,
tracker)
ret = Dict()
for dist in setdiff(keys(type_tracker.indices), [:Missing, :Observed])
summary_missing_only = provide(Diagnostics{Any}(),
reference = truth_matrix[tracker[type_conversion(Symbol, dist)][:Missing]],
input_data = predicted_matrix[tracker[type_conversion(Symbol, dist)][:Missing]])
summary_all = provide(Diagnostics{Any}(),
reference = truth_matrix[type_tracker[type_conversion(Symbol, dist)]],
input_data = predicted_matrix[type_tracker[type_conversion(Symbol, dist)]])
ret[dist] = Dict()
ret[dist][:MissingOnly] = summary_missing_only
ret[dist][:All] = summary_all
end
return ret
end
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 1687 | # early exit
# if (max(primfeas, dualfeas) < stoptol) | (iter == maxiter)
# breakyes = 1
# end
# if (max(primfeas, dualfeas) < sqrt(stoptol)) & (dualfeas > 1.5 * minimum(runhist.dualfeas[max(iter - 49, 1):iter])) & (iter > 150)
# breakyes = 2
# end
# tune ρ
# if (ρReset > 0) & (rem(iter, 10)==0)
# if (primfeas < 0.5 * dualfeas)
# ρ = 0.7 * ρ
# elseif (primfeas > 2 * dualfeas)
# ρ = 1.3 * ρ
# end
# end
# if (breakyes > 0)
# @printf("\n break = %1.0f\n", breakyes)
# break;
# end
# function sdpProjection(mat::Array{Float64, 2})
# λ, X = eigs(KrylovMethods(), mat, nev = 20)
# # @show(λ)
# # @show(X)
# # @show(size(λ))
# # @show(size(X))
# return project(λ, X)
# # posEigenValuesIndex = findall(x -> x > 0, λ);
# # posEigenValues = λ[posEigenValuesIndex];
# # posEigenVectors = X[:,posEigenValuesIndex];
# # projectedMatrix = posEigenVectors * diagm(0 => posEigenValues) *posEigenVectors';
# # return projectedMatrix;
# end
# function sdpProjection0(data)
# eigDecomposition = eigen(data);
# posEigenValuesIndex = findall(x -> x>0,eigDecomposition.values);
# posEigenValues = eigDecomposition.values[posEigenValuesIndex];
# posEigenVectors = eigDecomposition.vectors[:,posEigenValuesIndex];
# projectedMatrix = posEigenVectors * diagm(0 => posEigenValues) *posEigenVectors';
# return projectedMatrix;
# end
# # function logisticLoss(x,y)
# # f_x = Losses.σ.(x);
# # return -sum(y .* log.(f_x) + (1 .- y) .* log.(1 .- f_x));
# # end
# return X * diagm(0 => Λ) * X'
# TODO: further extract positive eigen values
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 25193 | module ADMM
using Printf
using LinearAlgebra
using SparseArrays
using ..Concepts
using ..Estimator
using ..ModelFitting
using ..Utilities
using ..Utilities.FastEigen
using ..Utilities.PrettyPrinter
using ..Losses
using ..MathLib
using Logging
import StatsBase
import LinearAlgebra:norm
# export complete
mutable struct RunHistory
primfeas::Array{Float64,1}
dualfeas::Array{Float64,1}
function RunHistory()
new(Array{Float64,1}(),Array{Float64,1}())
end
end
import KrylovKit
# function project(v,e)
# return e * diagm(0 => v) * e';
# end
const header_list = ["Iter",
" R(dual)",
"R(primal)",
"ℒ(Gaussian)",
"ℒ(Bernoulli)",
"ℒ(Poisson)",
"ℒ(NegBin) ",
" ℒ(Gamma) ",
"λ‖diag(Z)‖ᵢ",
" μ⟨I, X⟩ ",
" ‖Z₁₂‖ᵢ "]
function l1BallProjection(v,b)
if (norm(v,1) <= b);
return v
end
n = length(v);
nvec = hcat(1:n...)'[:];
vv = sort(abs.(v),lt = (x,y) -> !isless(x,y));
idxsort = sortperm(abs.(v),lt = (x,y) -> !isless(x,y));
vsum = cumsum(vv);
tmp = vv .-(vsum .- b)./nvec;
idx = findall(x->x>0,tmp);
if !isempty(idx);
k = maximum(idx);
else
println("something is wrong")
end
lam = (vsum[k] .- b) ./ k;
xx = zeros(length(idxsort));
xx[idxsort,1] = max.(vv .- lam,0);
x = sign.(v).*xx;
return x;
end
@private
function initialize_warmup(tracker, A)
warmup = Dict{Symbol, Array{Float64}}()
if haskey(tracker, :Bernoulli) && length(tracker[:Bernoulli][:Observed]) > 0
warmup[:Bernoulli] = rand(length(tracker[:Bernoulli][:Observed]), 1)
end
if haskey(tracker, :Gaussian) && length(tracker[:Gaussian][:Observed]) > 0
warmup[:Gaussian] = rand(length(tracker[:Gaussian][:Observed]), 1)
end
if haskey(tracker, :Poisson) && length(tracker[:Poisson][:Observed]) > 0
warmup[:Poisson] = rand(length(tracker[:Poisson][:Observed]), 1)
end
if haskey(tracker, :Gamma) && length(tracker[:Gamma][:Observed]) > 0
warmup[:Gamma] = rand(length(tracker[:Gamma][:Observed]), 1)
end
if haskey(tracker, :NegativeBinomial) && length(tracker[:NegativeBinomial][:Observed]) > 0
warmup[:NegativeBinomial] = rand(length(tracker[:NegativeBinomial][:Observed]), 1)
end
return warmup
end
@private
function ensure_feasible_data_type(set_of_types)
@info(set_of_types)
if !issubset(set_of_types, Set([:Gaussian,
:Poisson,
:NegativeBinomial,
:Gamma,
:Bernoulli]))
@error("Unrecognized data type.")
return nothing
end
end
@private
function ensure_valid_param_estimates(::Type{Val{:Gaussian}}, value::T) where T
return true
end
@private
function ensure_valid_param_estimates(::Type{Val{:Bernoulli}}, value::T) where T
return true
end
@private
function ensure_valid_param_estimates(::Type{Val{:Gamma}}, value::T) where T
return true
end
@private
function ensure_valid_param_estimates(::Type{Val{:Posson}}, value::T) where T
return true
end
@private
function ensure_valid_param_estimates(::Type{Val{:NegativeBinomial}}, value::T) where T
if keys(value) != Set((:p, :r))
@error("Unrecognized parameter name for Negative Binomial. Expected (:p, :r)")
return false
end
if !(0 < value[:p] < 1)
@error("Estimator for p in Negative Binomial distribution should be in (0, 1)")
return false
end
if value[:r] < 0
@error("Estimator for r in Negative Binomial distribution should be in (0, ∞)")
return false
end
return true
end
@private
function preprocess_user_input_estimators(input::Optional{Dict{Symbol, Dict{Symbol, T}}}) where T<:Real
if isnothing(input)
return Dict{Symbol, Any}()
end
ensure_feasible_data_type(keys(input))
processed_input = Dict{Symbol, Dict{Symbol, Any}}()
for dist in [:NegativeBinomial, :Gaussian, :Poisson, :Bernoulli, :Gamma]
if haskey(input, dist)
# @show(dist)
processed_input[dist] = Dict{Symbol, Any}()
processed_input[dist] = merge(processed_input[dist],
preprocess_user_input_param_estimates(Val{dist}, input[dist]))
end
end
return processed_input
end
@private
function preprocess_user_input_param_estimates(::Type{T}, value::S) where {T, S}
ensure_valid_param_estimates(T, value)
return Dict{Symbol, Any}(:user_input => value)
end
@private
function initialize_estimators(tracker, A, user_input_estimators)
estimators = Dict{Symbol, Dict{Symbol, Any}}()
for sym in [:Gaussian, :Bernoulli, :Poisson, :Gamma, :NegativeBinomial]
estimators[sym] = Dict{Symbol, Any}()
end
if haskey(tracker, :NegativeBinomial) && length(tracker[:NegativeBinomial][:Observed]) > 0
@info("Found negative binomial items. Use MOM for r and p")
estimators[:NegativeBinomial][:MOM] = Concepts.estimator(MOM{AbstractNegativeBinomial}(),
convert(Array{Float64},
A[tracker[:NegativeBinomial][:Observed]]))
@info(estimators[:NegativeBinomial][:MOM])
end
return merge(estimators, preprocess_user_input_estimators(user_input_estimators))
end
@private
function update(::Type{Val{:Gaussian}},
A ::Array{MaybeMissing{Float64}},
Y12 ::Array{Float64},
tracker,
ρ ::Float64,
gd_iter ::Int64,
warmup ::Dict{Symbol, Array{Float64}},
γ ::Float64,
use_autodiff ::Bool,
closed_form ::Bool)
if closed_form == true
if haskey(tracker, :Gaussian) && length(tracker[:Gaussian][:Observed]) > 0
Y12[tracker[:Gaussian][:Observed]] .= (1 / (1 + ρ)) * (A[tracker[:Gaussian][:Observed]] + ρ * Y12[tracker[:Gaussian][:Observed]])
end
else
if haskey(tracker, :Gaussian) && length(tracker[:Gaussian][:Observed]) > 0
Y12[tracker[:Gaussian][:Observed]] = train(use_autodiff ? provide(Loss{AbstractGaussian}()) : Loss{AbstractGaussian}(),
fx = warmup[:Gaussian],
y = A[tracker[:Gaussian][:Observed]],
c = Y12[tracker[:Gaussian][:Observed]],
ρ = ρ,
iter = gd_iter,
γ = 0.2)
warmup[:Gaussian] = Y12[tracker[:Gaussian][:Observed]]
end
end
end
@private
function update(::Type{Val{:Bernoulli}},
A ::Array{MaybeMissing{Float64}},
Y12 ::Array{Float64},
tracker,
ρ ::Float64,
gd_iter ::Int64,
warmup ::Dict{Symbol, Array{Float64}},
γ ::Float64,
use_autodiff ::Bool)
if haskey(tracker, :Bernoulli) && length(tracker[:Bernoulli][:Observed]) > 0
Y12[tracker[:Bernoulli][:Observed]] = train(use_autodiff ? provide(Loss{AbstractBernoulli}()) : Loss{AbstractBernoulli}(),
fx = warmup[:Bernoulli],
y = A[tracker[:Bernoulli][:Observed]],
c = Y12[tracker[:Bernoulli][:Observed]],
ρ = ρ,
iter = gd_iter,
γ = 0.2)
warmup[:Bernoulli] = Y12[tracker[:Bernoulli][:Observed]]
end
end
@private
function update(::Type{Val{:Poisson}},
A ::Array{MaybeMissing{Float64}},
Y12 ::Array{Float64},
tracker,
ρ ::Float64,
gd_iter ::Int64,
warmup ::Dict{Symbol, Array{Float64}},
γ ::Float64,
use_autodiff ::Bool)
if haskey(tracker, :Poisson) && length(tracker[:Poisson][:Observed]) > 0
Y12[tracker[:Poisson][:Observed]] = train(use_autodiff ? provide(Loss{AbstractPoisson}()) : Loss{AbstractPoisson}(),
fx = warmup[:Poisson],
y = A[tracker[:Poisson][:Observed]],
c = Y12[tracker[:Poisson][:Observed]],
ρ = ρ,
iter = gd_iter,
γ = 0.1)
warmup[:Poisson] = Y12[tracker[:Poisson][:Observed]]
end
end
@private
function update(::Type{Val{:Gamma}},
A ::Array{MaybeMissing{Float64}},
Y12 ::Array{Float64},
tracker,
ρ ::Float64,
gd_iter ::Int64,
warmup ::Dict{Symbol, Array{Float64}},
γ ::Float64, use_autodiff::Bool)
if haskey(tracker, :Gamma) && length(tracker[:Gamma][:Observed]) > 0
Y12[tracker[:Gamma][:Observed]] = train(use_autodiff ? provide(Loss{AbstractGamma}()) : Loss{AbstractGamma}(),
fx = warmup[:Gamma],
y = A[tracker[:Gamma][:Observed]],
c = Y12[tracker[:Gamma][:Observed]],
ρ = ρ,
iter = gd_iter,
γ = 0.2)
warmup[:Gamma] = Y12[tracker[:Gamma][:Observed]]
end
end
@private
function update(::Type{Val{:NegativeBinomial}},
A ::Array{MaybeMissing{Float64}},
Y12 ::Array{Float64},
tracker,
ρ ::Float64,
gd_iter ::Int64,
warmup ::Dict{Symbol, Array{Float64}},
γ ::Float64,
use_autodiff ::Bool,
estimator ::Dict{Symbol, Any})
if haskey(tracker, :NegativeBinomial) && length(tracker[:NegativeBinomial][:Observed]) > 0
local r_est = nothing
if haskey(estimator, :user_input)
r_est = estimator[:user_input][:r]
else
r_est = estimator[:MOM][:r]
end
Y12[tracker[:NegativeBinomial][:Observed]] = negative_binomial_train(fx = warmup[:NegativeBinomial],
y = A[tracker[:NegativeBinomial][:Observed]],
c = Y12[tracker[:NegativeBinomial][:Observed]],
ρ = ρ,
iter = gd_iter,
γ = 0.2,
r_estimate = r_est)
warmup[:NegativeBinomial] = Y12[tracker[:NegativeBinomial][:Observed]]
end
end
@private
function update(::Type{Val{:ρ}}, primal_feasibility::Float64, dual_feasibility::Float64, current::Float64 = nothing)
if (primal_feasibility < 0.5 * dual_feasibility)
return 0.7 * current
elseif (primal_feasibility > 2 * dual_feasibility)
return 1.3 * current
end
return current
end
@private
function balance_gap(ρ::Float64, primal_feasibility::Float64, dual_feasibility::Float64)
if (primal_feasibility < 0.5 * dual_feasibility)
return 0.7 * ρ
elseif (primal_feasibility > 2 * dual_feasibility)
return 1.3 * ρ
end
return ρ
end
# update
const update(arg::Symbol, args...) = update(Val{arg}, args...)
@private
function calculate_primal_and_dual_residual(X, Z, W, C, Xinput, ρ)
Fnorm = x -> norm(x,2);
Xdual = -ρ * (X - Xinput)
Zdual = ρ * (Z - C)
normX = 1 + Fnorm(X)
primfeas = Fnorm(X - Z) / normX
err1 = 1/ρ * Fnorm(W - Xdual)
err2 = 1/ρ * Fnorm(W - Zdual)
dualfeas = maximum([err1, err2]) / normX
return primfeas, dualfeas
end
function set_diagonal(mat::Array{Float64, 2}, val::Array{Float64, 1})
[mat[i, i] = val[i] for i in 1:length(val)]
end
function calculate_Z12_update(A, C,tracker, ρ, α, warmup, use_autodiff, gd_iter, estimators, closed_form)
d1, d2 = size(A)
Z12 = C[1:d1, (d1+1):(d1+d2)]
update(:Gaussian, A, Z12, tracker, ρ, gd_iter, warmup, 0.2, use_autodiff, closed_form)
update(:Bernoulli, A, Z12, tracker, ρ, gd_iter, warmup, 0.2, use_autodiff)
update(:Poisson, A, Z12, tracker, ρ, gd_iter, warmup, 0.2, use_autodiff)
update(:Gamma, A, Z12, tracker, ρ, gd_iter, warmup, 0.005, use_autodiff)
update(:NegativeBinomial, A, Z12, tracker, ρ, gd_iter, warmup, 0.005, use_autodiff, estimators[:NegativeBinomial])
project!(ClosedInterval{Float64}(-α, α), Z12)
return Z12
end
function set_block_12(mat, d1, d2, val)
@. mat[1:d1, (d1+1):(d1+d2)] = val
end
function set_block_21(mat, d1, d2, val)
@. mat[(d1+1):(d1+d2), 1:d1] = val
end
@private
function initialize_trackers(A::Array{MaybeMissing{Float64}}, type_assignment)
type_tracker = Utilities.IndexTracker{Symbol}()
if type_assignment == nothing
disjoint_join(type_tracker, Concepts.fit(DistributionDeduction(), A))
else
disjoint_join(type_tracker, type_assignment)
end
disjoint_join(type_tracker, Concepts.fit(ObservedOrMissing(), A))
return groupby(type_tracker, [:Observed, :Missing]), type_tracker
end
@private
function ensure_feasible(A::Array{MaybeMissing{Float64}})
if isnothing(A)
@error(io, "please provide data matrix")
throw(MethodError())
end
end
function format_log_data(x)
if x == -10000000
return "N/A"
end
return @sprintf("%3.2e", x)
end
@private
function print_optimization_log(iter,A, X, Z, Z12, W, II, Rp, Rd, ρ, λ, μ, tracker, estimators, io)
R = abs.(maximum(diag(Z)))
local gaussian_loss = -10000000
local bernoulli_loss = -10000000
local poisson_loss = -10000000
local gamma_loss = -10000000
local negative_binomial_loss = -10000000
if haskey(tracker, :Gaussian)
# gaussian_loss = norm(Z12[tracker[:Gaussian][:Observed]] - A[tracker[:Gaussian][:Observed]])^2
gaussian_loss = evaluate(Loss{AbstractGaussian}(),
Z12[tracker[:Gaussian][:Observed]],
A[tracker[:Gaussian][:Observed]],
similar(Z12[tracker[:Gaussian][:Observed]]),
0)
end
if haskey(tracker, :Bernoulli)
bernoulli_loss = evaluate(Loss{AbstractBernoulli}(),
Z12[tracker[:Bernoulli][:Observed]],
A[tracker[:Bernoulli][:Observed]],
similar(Z12[tracker[:Bernoulli][:Observed]]),
0)
end
if haskey(tracker, :NegativeBinomial)
local r_est = nothing
if haskey(estimators[:NegativeBinomial], :user_input)
r_est = estimators[:NegativeBinomial][:user_input][:r]
else
r_est = estimators[:NegativeBinomial][:MOM][:r]
end
negative_binomial_loss = evaluate(Loss{AbstractNegativeBinomial}(),
Z12[tracker[:NegativeBinomial][:Observed]],
A[tracker[:NegativeBinomial][:Observed]],
similar(Z12[tracker[:NegativeBinomial][:Observed]]),
0,
r_estimate = r_est)
end
if haskey(tracker, :Poisson)
poisson_loss = evaluate(Loss{AbstractPoisson}(),
Z12[tracker[:Poisson][:Observed]],
A[tracker[:Poisson][:Observed]],
similar(Z12[tracker[:Poisson][:Observed]]),
0)
end
if haskey(tracker, :Gamma)
gamma_loss = evaluate(Loss{AbstractGamma}(),
Z12[tracker[:Gamma][:Observed]],
A[tracker[:Gamma][:Observed]],
similar(Z12[tracker[:Gamma][:Observed]]),
0)
# gamma_loss = 0
end
data = [iter,
Rp,
Rd,
gaussian_loss,
bernoulli_loss,
poisson_loss,
negative_binomial_loss,
gamma_loss,
maximum(abs.(Z12)),
μ * tr(II * X),
abs.(maximum(diag(Z)))
]
new_data = map(x -> format_log_data(x) ,data)
new_data[1] = string(iter)
add_row(header_list, data=new_data, io = io)
end
#TODO
# function standardize(A, tracker, estimators)
# A[tracker[:Gaussian][:Observed]] .= (A[tracker[:Gaussian][:Observed]] .- estimators[:Gaussian][:μ]) ./ estimators[:Gaussian][:σ]
# end
#TODO
# function destandardize(A, type_tracker, estimators)
# A[tracker[:Gaussian]]] .= (A[tracker] .* estimators[:Gaussian][:σ]) .+ estimators[:Gaussian][:μ]
# end
struct OneShotADMM end
function Concepts.complete(model::OneShotADMM;
A::Array{MaybeMissing{Float64}} = nothing,
α::Float64 = maximum(A[findall(x -> !ismissing(x),A)]),
λ::Float64 = 5e-1,
μ::Float64 = 5e-4,
ρ::Float64 = 0.3,
τ::Float64 = 1.618,
maxiter::Int64 = 200,
stoptol::Float64 = 1e-5,
use_autodiff::Bool = false,
gd_iter::Int64 = 50,
debug_mode::Bool = false,
interactive_plot = false,
type_assignment = nothing,
warmup = nothing,
start_var = nothing,
dynamic_ρ = true,
user_input_estimators = nothing,
project_rank = nothing,
io::IO = Base.stdout,
eigen_solver = KrylovMethods(),
closed_form_update = false)
return complete(;A = A,
α = α,
λ = λ,
μ = μ,
ρ = ρ,
τ = τ,
maxiter = maxiter,
stoptol = stoptol,
use_autodiff = use_autodiff,
gd_iter = gd_iter,
debug_mode = debug_mode,
interactive_plot = interactive_plot,
type_assignment = type_assignment,
warmup = warmup,
start_var = start_var,
dynamic_ρ,
user_input_estimators = user_input_estimators,
project_rank = project_rank,
io = io,
eigen_solver = eigen_solver,
closed_form_update = closed_form_update)
end
function Concepts.complete(;A::Array{MaybeMissing{Float64}} = nothing,
α::Float64 = maximum(A[findall(x -> !ismissing(x),A)]),
λ::Float64 = 5e-1,
μ::Float64 = 5e-4,
ρ::Float64 = 0.3,
τ::Float64 = 1.618,
maxiter::Int64 = 200,
stoptol::Float64 = 1e-5,
use_autodiff::Bool = false,
gd_iter::Int64 = 50,
debug_mode::Bool = false,
interactive_plot = false,
type_assignment = nothing,
warmup = nothing,
start_var = nothing,
dynamic_ρ = true,
user_input_estimators = nothing,
project_rank = nothing,
io::IO = Base.stdout,
eigen_solver = KrylovMethods(),
closed_form_update = false)
logger = SimpleLogger(io)
global_logger(logger)
if isnothing(project_rank)
@info("Using Full Eigen Decomposition.")
else
@info("Using Fast Eigen")
end
ensure_feasible(A)
d1, d2 = size(A);
Z::Array{Float64, 2} = zeros(d1 + d2, d1 + d2)
X::Array{Float64, 2} = zeros(d1 + d2, d1 + d2)
W::Array{Float64, 2} = zeros(d1 + d2, d1 + d2)
C::Array{Float64, 2} = zeros(d1 + d2, d1 + d2)
if !isnothing(start_var)
Z = start_var[:Z]
X = start_var[:X]
W = start_var[:W]
C = start_var[:C]
end
Xinput::Array{Float64, 2} = zeros(d1 + d2, d1 + d2)
II = sparse(1.0I, d1 + d2, d1 + d2)
tracker, type_tracker = initialize_trackers(A, type_assignment)
# initialize warmup input for various gradient descent procedures
if isnothing(warmup)
warmup::Dict{Symbol, Array{Float64}} = initialize_warmup(tracker, A)
end
# initialize various estimators
estimators::Dict{Symbol, Any} = initialize_estimators(tracker, A, user_input_estimators)
# print optimization path table header
table_header(header_list, io = io)
for iter = 1:maxiter
@. Xinput = Z + W/ρ
# step 1
# try
if isnothing(project_rank)
X = project(SemidefiniteCone(), Z + W / ρ - (μ / ρ) * II)
else
X = project(SemidefiniteCone(rank = project_rank), Z + W / ρ - (μ / ρ) * II,
eigs_implementation = eigen_solver)
end
# catch
# @warn("Manual fix for numerical instability")
# fix = Z + W / ρ - (μ / ρ) * II
# fix[findall(x -> x== Inf || isnan(x), fix)] .= rand()
# X = project(SemidefiniteCone(rank = 20), fix)
# end
# Step 2
@. C = X - 1/ρ * W; @. Z = C
Z12 = calculate_Z12_update(A, C, tracker, ρ, α, warmup, use_autodiff, gd_iter, estimators, closed_form_update)
set_block_12(Z, d1, d2, Z12)
set_block_21(Z, d1, d2, Z12')
set_diagonal(Z, diag(C) - (λ / ρ) * l1BallProjection(diag(C) * ρ / λ, 1))
# step 3
@. W = W + τ * ρ * (Z - X)
if rem(iter, 10)==1
primfeas, dualfeas = calculate_primal_and_dual_residual(X, Z, W, C, Xinput, ρ)
print_optimization_log(iter, A, X, Z, Z12, W, II, primfeas, dualfeas, ρ, λ, μ, tracker, estimators, io)
if dynamic_ρ
ρ = balance_gap(ρ, primfeas, dualfeas)
end
end
end
completedMatrix = C[1:d1, (d1+1):(d1+d2)]
last_info = Dict(:Z => Z,
:X => X,
:W => W,
:C => C,
:warmup => warmup)
return completedMatrix, type_tracker, tracker, last_info
end
include("./ChainedADMM.jl")
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 6266 | struct ChainedADMM end
function sample_without_replacement!(collection, n::Int64) where T<:Any
sample = []
for i in 1:n
push!(sample, splice!(collection,rand(eachindex(collection))))
end
return sample
end
function initialize_chained_trackers(A::Array{MaybeMissing{Float64}}, type_assignment)
type_tracker = Utilities.IndexTracker{Symbol}()
if type_assignment == nothing
Concepts.disjoint_join(type_tracker, Concepts.fit(DistributionDeduction(), A))
else
Concepts.disjoint_join(type_tracker, type_assignment)
end
Concepts.disjoint_join(type_tracker, Concepts.fit(ObservedOrMissing(), A))
result = Concepts.groupby(type_tracker, [:Observed, :Missing])
result[:Observed][:Total] = Base.findall(!ismissing, A)
result[:Missing][:Total] = Base.findall(ismissing, A)
result[:SingleView] = type_tracker
return result
end
function Concepts.complete(model::ChainedADMM;
block_size = nothing,
imputation_round::Union{Int64, Nothing} = 10,
A::Array{MaybeMissing{Float64}} = nothing,
α::Float64 = maximum(A[findall(x -> !ismissing(x),A)]),
λ::Float64 = 5e-1,
μ::Float64 = 5e-4,
ρ::Float64 = 0.3,
τ::Float64 = 1.618,
maxiter::Int64 = 200,
stoptol::Float64 = 1e-5,
use_autodiff::Bool = false,
gd_iter::Int64 = 50,
debug_mode::Bool = false,
interactive_plot = false,
type_assignment = nothing,
warmup = nothing,
dynamic_ρ = true,
user_input_estimators = nothing,
project_rank = nothing,
io::IO = Base.stdout,
eigen_solver = KrylovMethods(),
closed_form_update = false)
o_tracker, o_type_tracker = initialize_trackers(A, type_assignment)
tracker = initialize_chained_trackers(A, type_assignment)
missing_entries = deepcopy(tracker[:Missing][:Total])
# @show(missing_entries[1:10])
observed_entries = deepcopy(tracker[:Observed][:Total])
# @show(observed_entries[1:10])
if !isnothing(imputation_round)
block_size = trunc(Int64, Base.ceil(length(missing_entries) / imputation_round))
@info @sprintf("imputation round enforced, current block size is %d.\n", block_size)
end
# @show(imputation_round)
if isnothing(warmup)
warmup::Dict{Symbol, Array{Float64}} = initialize_warmup(tracker, A)
end
imputed = A
round = 0
local result_completed_matrix = nothing
local result_type_tracker = nothing
local result_tracker = nothing
local result_last_var = nothing
while length(missing_entries) > 0
round = round + 1
@info(round)
block_samples = sample_without_replacement!(missing_entries, min(block_size, length(missing_entries)))
result_completed_matrix, result_type_tracker, result_tracker, result_last_var = complete(A = imputed,
α = α,
λ = λ,
μ = μ,
ρ = ρ,
τ = τ,
maxiter = maxiter,
stoptol = stoptol,
use_autodiff = use_autodiff,
gd_iter = gd_iter,
start_var = result_last_var,
debug_mode = debug_mode,
interactive_plot = interactive_plot,
type_assignment = type_assignment,
dynamic_ρ = dynamic_ρ,
user_input_estimators = user_input_estimators,
project_rank = project_rank,
io = io,
eigen_solver = eigen_solver,
closed_form_update = closed_form_update)
predicted_matrix = predict(MatrixCompletionModel(),
completed_matrix = result_completed_matrix,
type_tracker = result_type_tracker)
# @show("update imputed")
for index in block_samples
imputed[index] = predicted_matrix[index]
end
# if haskey(tracker, :Bernoulli) && length(tracker[:Bernoulli][:Observed]) > 0
# if haskey(tracker, :Bernoulli) && length(tracker[:Bernoulli][:Observed]) > 0
# warmup[:Bernoulli] =
# end
end
return result_completed_matrix, o_type_tracker, o_tracker, imputed
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 52 | include("./ADMM.jl")
# include("./ChainedADMM.jl")
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 648 | module MathLib
import LinearAlgebra
macro api(as,of)
eval(Expr(:toplevel,:($as=$of),:(export $as)))
end
using ..MathLibSignatures
using ..Concepts
@api MathematicalObject MathLibSignatures.MathematicalObject,
@api Cone MathLibSignatures.Cone
@api Interval MathLibSignatures.Interval
@api SemidefiniteCone MathLibSignatures.SemidefiniteCone
@api ClosedInterval MathLibSignatures.ClosedInterval
@api project MathLibSignatures.project
@api project! MathLibSignatures.project!
function LinearAlgebra.rank(obj::SemidefiniteCone)
return obj.rank
end
include("./Projections.jl")
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 1058 | module MathLibSignatures
using ..Concepts
export MathematicalObject,
Cone,
Interval,
SemidefiniteCone,
ClosedInterval
abstract type MathematicalObject end
abstract type Cone <: MathematicalObject end
abstract type Interval <: MathematicalObject end
struct SemidefiniteCone <: Cone
rank::Optional{Int64}
function SemidefiniteCone(; rank::Optional{Int64} = nothing)
if rank == nothing
return new(nothing)
end
if rank <= 0
@warn("Rank should never be below zero. Check again.")
throw(MethodError())
end
return new(rank)
end
end
struct ClosedInterval{T} <: Interval where T<:Real
ll::Optional{T}
rr::Optional{T}
function ClosedInterval{T}(ll::T, rr::T) where T<:Real
if ll > rr
@warn("Right end point of the interval shoud be bigger than left, return an empty interval instead.")
return new{T}(nothing, nothing)
end
return new{T}(ll, rr)
end
end
function project() end
function project!() end
function scope_test()
@info("inside MathLib.Concepts")
end
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 1199 | using .MathLibSignatures
using ..Utilities.FastEigen
using LinearAlgebra
function MathLibSignatures.project(to::SemidefiniteCone, mat::Array{Float64, 2};
eigs_implementation = KrylovMethods())
if isnothing(rank(to))
# do a full projection
# @warn("Doing full eigen projection, could be costly!")
eigDecomposition = eigen(mat);
posEigenValuesIndex = findall(x -> real(x) > 0,eigDecomposition.values);
posEigenValues = eigDecomposition.values[posEigenValuesIndex];
posEigenVectors = eigDecomposition.vectors[:,posEigenValuesIndex];
projectedMatrix = posEigenVectors * diagm(0 => posEigenValues) *posEigenVectors';
return projectedMatrix;
end
# we are computing the full projection
Λ, X = eigs(eigs_implementation, mat, nev = to.rank)
# return X * diagm(0 => Λ) * X'
id = findall(x -> real(x) > 0, Λ)
return X[:, id] * diagm(0 => Λ[id]) * (X[:,id])'
end
function MathLibSignatures.project(to::ClosedInterval, x::AutoboxedArray{Float64})
return max.(to.ll, min.(x, to.rr))
end
function MathLibSignatures.project!(to::ClosedInterval, x::Array{Float64})
@. x = max.(to.ll, min.(x, to.rr))
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 6429 | include("./lowrankmodels/LowRankModels.jl")
module ALM
using Printf
import ..Concepts
import ..Concepts: MaybeMissing, type_conversion
import ..Utilities
import ..LowRankModels
import ..ModelFitting.ObservedOrMissing
using Logging
export ChainedALM, OneShotALM
struct GLRMLosses end
struct ChainedALM end
struct OneShotALM end
struct MeanImputation end
struct ChainedEquations end
struct ChainedTruncatedSVD end
function sample_without_replacement!(collection, n::Int64) where T<:Any
sample = []
for i in 1:n
push!(sample, splice!(collection,rand(eachindex(collection))))
end
return sample
end
function update_observed_entries!(observed_entries, sampled_entries)
for index in sampled_entries
Base.push!(observed_entries, index)
end
end
function update_imputed_entries!(cur_imputed, sampled_entries, X, Y)
XtY = X' * Y
for index in sampled_entries
cur_imputed[index] = XtY[index]
end
end
function Concepts.type_conversion(::Type{GLRMLosses}, x::Symbol)
if x == :Gaussian
return LowRankModels.QuadLoss()
else
return nothing
end
end
function initialize_trackers(A::Array{MaybeMissing{Float64}}, type_assignment)
type_tracker = Utilities.IndexTracker{Symbol}()
if type_assignment == nothing
Concepts.disjoint_join(type_tracker, Concepts.fit(DistributionDeduction(), A))
else
Concepts.disjoint_join(type_tracker, type_assignment)
end
Concepts.disjoint_join(type_tracker, Concepts.fit(ObservedOrMissing(), A))
result = Concepts.groupby(type_tracker, [:Observed, :Missing])
result[:Observed][:Total] = Base.findall(!ismissing, A)
result[:Missing][:Total] = Base.findall(ismissing, A)
result[:SingleView] = type_tracker
return result
end
function prepare_loss_functions(type_assignment)
return Base.map(x -> type_conversion(GLRMLosses ,x), type_assignment[1, :])
end
function Concepts.complete(model::OneShotALM;
A::Array{MaybeMissing{Float64}},
type_assignment,
rx = LowRankModels.QuadReg(0),
ry = LowRankModels.QuadReg(0),
target_rank,
initialX = nothing,
initialY = nothing,
proximal_params = nothing)
row, col = size(A)
tracker = initialize_trackers(A, type_assignment)
missing_entries = deepcopy(tracker[:Missing][:Total])
observed_entries = deepcopy(tracker[:Observed][:Total])
loss = prepare_loss_functions(type_assignment)
imputed = A
@info @sprintf("total number of entries: %d\n", row * col)
@info @sprintf("total number of missing entries: %d (%.4f%%)\n", length(missing_entries), length(missing_entries) / (row * col))
@info @sprintf("target rank: %d\n", target_rank)
if isnothing(initialX)
@info "initial state of X in undeteced, using randomized initilization"
initialX = randn(target_rank, row)
end
if isnothing(initialY)
@info "initial state of Y in undeteced, using randomized initilization"
initialY = randn(target_rank, col)
end
glrm = LowRankModels.GLRM(imputed, loss, rx, ry, target_rank, obs = observed_entries, X = initialX, Y = initialY);
local X, Y, ch
if isnothing(proximal_params)
X, Y, ch = LowRankModels.fit!(glrm)
else
X, Y, ch = LowRankModels.fit!(glrm, proximal_params)
end
update_imputed_entries!(imputed, missing_entries, X, Y)
return imputed, X, Y, tracker
end
function Concepts.complete(model::ChainedALM;
A::Array{MaybeMissing{Float64}},
type_assignment,
block_size,
imputation_round::Union{Int64, Nothing} = nothing,
rx = LowRankModels.QuadReg(0),
ry = LowRankModels.QuadReg(0),
target_rank,
initialX = nothing,
initialY = nothing,
proximal_params = nothing)
row, col = size(A)
tracker = initialize_trackers(A, type_assignment)
missing_entries = deepcopy(tracker[:Missing][:Total])
observed_entries = deepcopy(tracker[:Observed][:Total])
if !isnothing(imputation_round)
block_size = trunc(Int64, Base.ceil(length(missing_entries) / imputation_round))
@info @sprintf("imputation round enforced, current block size is %d.\n", block_size)
end
@info @sprintf("total number of entries: %d\n", row * col)
@info @sprintf("total number of missing entries: %d (%.4f%%)\n", length(missing_entries), length(missing_entries) / (row * col))
@info @sprintf("target rank: %d\n", target_rank)
@info @sprintf("block size: %d\n", block_size)
@info @sprintf("rounds of completion expected: %d\n", Base.ceil(length(missing_entries) / block_size))
if isnothing(initialX)
@info "initial state of X in undeteced, using randomized initilization"
initialX = randn(target_rank, row)
end
if isnothing(initialY)
@info "initial state of Y in undeteced, using randomized initilization"
initialY = randn(target_rank, col)
end
imputed = A
warmup = false
prevX = []; prevY=[];
loss = prepare_loss_functions(type_assignment)
warmup = false
round = 0
while length(missing_entries) > 0
round += 1
println(round)
block_samples = sample_without_replacement!(missing_entries, min(block_size, length(missing_entries)))
if (warmup == false)
glrm = LowRankModels.GLRM(imputed, loss, rx, ry, target_rank, obs = observed_entries, X = initialX, Y = initialY);
else
glrm = LowRankModels.GLRM(imputed, loss, rx, ry, target_rank, obs = observed_entries, X = prevX, Y = prevY)
end
warmup = true
local X, Y, ch
if isnothing(proximal_params)
X, Y, ch = LowRankModels.fit!(glrm)
else
X, Y, ch = LowRankModels.fit!(glrm, proximal_params)
end
prevX = X
prevY = Y
update_observed_entries!(observed_entries, block_samples)
update_imputed_entries!(imputed, block_samples, X, Y)
end
return imputed, prevX, prevY, tracker
end
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 1302 | # __precompile__()
module LowRankModels
using Printf
using SharedArrays
using SparseArrays
using Random
using Statistics
using DataFrames
import LinearAlgebra: dot, norm, Diagonal, rmul!, mul!
import Base: show
import StatsBase: fit!, mode, mean, var, std
# define losses, regularizers, convergence history
include("domains.jl")
include("losses.jl")
include("impute_and_err.jl")
include("regularizers.jl")
include("convergence.jl")
# define basic data type(s)
include("glrm.jl")
include("shareglrm.jl")
# modify models (eg scaling and offsets) and evaluate fit
include("modify_glrm.jl")
include("evaluate_fit.jl")
# fitting algorithms
include("fit.jl")
if Threads.nthreads() > 1
include("algorithms/proxgrad_multithread.jl")
else
include("algorithms/proxgrad.jl")
end
include("algorithms/sparse_proxgrad.jl")
include("algorithms/quad_streaming.jl")
# initialization methods
include("rsvd.jl")
include("initialize.jl")
# fancy fun on top of low rank models
include("simple_glrms.jl")
include("cross_validate.jl")
include("fit_dataframe.jl")
include("sample.jl")
# this takes to long to load for normal use
# include("plot.jl")
# utilities
include("utilities/conveniencemethods.jl")
include("utilities/deprecated.jl")
# ScikitLearn.jl compatibility
include("scikitlearn.jl")
end # module
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 1458 | export ConvergenceHistory, update_ch!
mutable struct ConvergenceHistory
name::AbstractString
objective::Array
dual_objective::Array
primal_residual::Array
dual_residual::Array
times::Array
stepsizes::Array
optval
end
ConvergenceHistory(name::AbstractString,optval=0) = ConvergenceHistory(name,Float64[],Float64[],Float64[],Float64[],Float64[],Float64[],optval)
ConvergenceHistory() = ConvergenceHistory("unnamed_convergence_history")
function update_ch!(ch::ConvergenceHistory, dt::Number, obj::Number,
stepsize::Number=0, pr::Number=0, dr::Number=0)
push!(ch.objective,obj)
push!(ch.primal_residual,pr)
push!(ch.dual_residual,dr)
push!(ch.stepsizes,stepsize)
if isempty(ch.times)
push!(ch.times,dt)
else
push!(ch.times,ch.times[end]+dt)
end
end
function update_ch!(ch::ConvergenceHistory, dt; obj=0, stepsize=0, pr=0, dr=0, dual_obj=0)
push!(ch.objective,obj)
push!(ch.dual_objective,dual_obj)
push!(ch.primal_residual,pr)
push!(ch.dual_residual,dr)
push!(ch.stepsizes,stepsize)
if isempty(ch.times)
push!(ch.times,dt)
else
push!(ch.times,ch.times[end]+dt)
end
end
function show(ch::ConvergenceHistory)
print("Convergence History for $(ch.name)\n\n")
@printf "%16s%16s\n" "time (s)" "objective"
for i=1:length(ch.objective)
@printf "%16.2e%16.4e\n" ch.times[i] ch.objective[i]
end
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 14717 | export cross_validate, cv_by_iter, regularization_path, get_train_and_test, precision_at_k
# the loss function evaluates the objective minus the regularization
# it is the default error metric
loss_fn(args...; kwargs...) = objective(args...; include_regularization=false, kwargs...)
# to use with error_metric when we have domains in the namespace, call as:
# cross_validate(glrm, error_fn = error_metric(glrm,domains,glrm.X,glrm.Y))
function cross_validate(glrm::AbstractGLRM;
nfolds=5, params=Params(),
verbose=true, use_folds=nfolds,
error_fn=loss_fn,
init=nothing,
do_obs_check = false)
if verbose println("flattening observations") end
# obs = flattenarray(map(ijs->map(j->(ijs[1],j),ijs[2]),zip(1:length(glrm.observed_features),glrm.observed_features)))
obs = flatten_observations(glrm.observed_features)
if verbose println("computing CV folds") end
folds = getfolds(obs, nfolds, size(glrm.A)..., do_check = do_obs_check)
train_glrms = Array{typeof(glrm)}(undef, nfolds)
test_glrms = Array{typeof(glrm)}(undef, nfolds)
train_error = Array{Float64}(undef, nfolds)
test_error = Array{Float64}(undef, nfolds)
for ifold=1:use_folds
if verbose println("\nforming train and test GLRM for fold $ifold") end
train_observed_features, train_observed_examples, test_observed_features, test_observed_examples = folds[ifold]
ntrain = sum(map(length, train_observed_features))
ntest = sum(map(length, test_observed_features))
if verbose println("training model on $ntrain samples and testing on $ntest") end
# form glrm on training dataset
train_glrms[ifold] = copy_estimate(glrm)
train_glrms[ifold].observed_examples = train_observed_examples
train_glrms[ifold].observed_features = train_observed_features
# form glrm on testing dataset
test_glrms[ifold] = copy_estimate(glrm)
test_glrms[ifold].observed_examples = test_observed_examples
test_glrms[ifold].observed_features = test_observed_features
# evaluate train and test error
if verbose println("fitting train GLRM for fold $ifold") end
if init != nothing
init(train_glrms[ifold])
end
fit!(train_glrms[ifold], params, verbose=verbose)
if verbose println("computing train and test error for fold $ifold:") end
train_error[ifold] = error_fn(train_glrms[ifold],
parameter_estimate(train_glrms[ifold])...) / ntrain
if verbose println("\ttrain error: $(train_error[ifold])") end
test_error[ifold] = error_fn(test_glrms[ifold],
parameter_estimate(train_glrms[ifold])...) / ntest
if verbose println("\ttest error: $(test_error[ifold])") end
end
return train_error, test_error, train_glrms, test_glrms
end
function getfolds(obs::Array{Tuple{Int,Int},1}, nfolds, m, n; ntrials = 5, do_check = true)
# partition elements of obs into nfolds groups
groups = Array{Int}(undef, size(obs))
rand!(groups, 1:nfolds) # fill an array with random 1 through N
# create the training and testing observations for each fold
folds = Array{Tuple}(undef, nfolds)
for itrial = 1:ntrials
enough_observations = 0
for ifold=1:nfolds
train = obs[filter(i->groups[i]!=ifold, 1:length(obs))] # all the obs that didn't get the ifold label
train_observed_features, train_observed_examples = sort_observations(train,m,n)
if !do_check ||
(check_enough_observations(train_observed_features) && check_enough_observations(train_observed_examples))
enough_observations += 1
else
@warn("Not enough data to cross validate; one of the cross validation folds has no observations in one row or column. Trying again...")
break
end
test = obs[filter(i->groups[i]==ifold, 1:length(obs))] # all the obs that did
test_observed_features, test_observed_examples = sort_observations(test,m,n,check_empty=false)
folds[ifold] = (train_observed_features, train_observed_examples,
test_observed_features, test_observed_examples)
end
if enough_observations == nfolds
return folds
end
end
error("Not enough data to cross validate automatically.")
end
function check_enough_observations(observed_examples_or_features)
all(map(length, observed_examples_or_features) .> 0)
end
function get_train_and_test(obs, m, n, holdout_proportion=.1)
# generate random uniform number for each observation
groups = Array{Float64}(undef, size(obs))
rand!(groups)
# create the training and testing observations
# observation is in test set if random number < holdout_proportion
train = obs[filter(i->(groups[i]>=holdout_proportion), 1:length(obs))]
train_observed_features, train_observed_examples = sort_observations(train,m,n)
test = obs[filter(i->(groups[i]<holdout_proportion), 1:length(obs))]
test_observed_features, test_observed_examples = sort_observations(test,m,n,check_empty=false)
return (train_observed_features, train_observed_examples,
test_observed_features, test_observed_examples)
end
function flatten_observations(observed_features::ObsArray)
obs = Array{Tuple{Int,Int}}(undef, 0)
for (i, features_in_example_i) in enumerate(observed_features)
for j in features_in_example_i
push!(obs, (i,j))
end
end
return obs
end
function flatten(x, y)
state = start(x)
if state==false
push!(y, x)
else
while !done(x, state)
(item, state) = next(x, state)
flatten(item, y)
end
end
y
end
flatten(x::Array{T}) where T=flatten(x,Array(T, 0))
function flattenarray(x, y)
if typeof(x)<:Array
for xi in x
flattenarray(xi, y)
end
else
push!(y, x)
end
y
end
flattenarray(x::Array{T}) where T=flattenarray(x,Array(T, 0))
function cv_by_iter(glrm::AbstractGLRM, holdout_proportion=.1,
params=Params(100,max_iter=1,abs_tol=.01,min_stepsize=.01),
ch = ConvergenceHistory("cv_by_iter");
verbose=true)
# obs = flattenarray(map(ijs->map(j->(ijs[1],j),ijs[2]),zip(1:length(glrm.observed_features),glrm.observed_features)))
obs = flatten_observations(glrm.observed_features)
train_observed_features, train_observed_examples, test_observed_features, test_observed_examples =
get_train_and_test(obs, size(glrm.A)..., holdout_proportion)
# form glrm on training dataset
train_glrm = copy_estimate(glrm)
train_glrm.observed_examples = train_observed_examples
train_glrm.observed_features = train_observed_features
# form glrm on testing dataset
test_glrm = copy_estimate(glrm)
test_glrm.observed_examples = test_observed_examples
test_glrm.observed_features = test_observed_features
ntrain = sum(map(length, train_glrm.observed_features))
ntest = sum(map(length, test_glrm.observed_features))
niters = params.max_iter
params.max_iter = 1
train_error = Array{Float64}(undef, niters)
test_error = Array{Float64}(undef, niters)
if verbose
@printf("%12s%12s%12s\n", "train error", "test error", "time")
t0 = time()
end
for iter=1:niters
# evaluate train and test error
fit!(train_glrm, params, ch=ch, verbose=false)
train_error[iter] = ch.objective[end] # objective(train_glrm, parameter_estimate(train_glrm)..., include_regularization=false)/ntrain
test_error[iter] = objective(test_glrm, parameter_estimate(train_glrm)..., include_regularization=false)/ntest
if verbose
@printf("%12.4e%12.4e%12.4e\n", train_error[iter], test_error[iter], time() - t0)
end
end
return train_error, test_error
end
function regularization_path(glrm::AbstractGLRM; params=Params(), reg_params=exp10.(range(2,stop=-2,length=5)),
holdout_proportion=.1, verbose=true,
ch::ConvergenceHistory=ConvergenceHistory("reg_path"))
if verbose println("flattening observations") end
# obs = flattenarray(map(ijs->map(j->(ijs[1],j),ijs[2]),zip(1:length(glrm.observed_features),glrm.observed_features)))
obs = flatten_observations(glrm.observed_features)
if verbose println("splitting train and test sets") end
train_observed_features, train_observed_examples, test_observed_features, test_observed_examples =
get_train_and_test(obs, size(glrm.A)..., holdout_proportion)
if verbose println("forming train and test GLRMs") end
# form glrm on training dataset
train_glrm = copy_estimate(glrm)
train_glrm.observed_examples = train_observed_examples
train_glrm.observed_features = train_observed_features
# form glrm on testing dataset
test_glrm = copy_estimate(glrm)
test_glrm.observed_examples = test_observed_examples
test_glrm.observed_features = test_observed_features
return regularization_path(train_glrm, test_glrm; params=params, reg_params=reg_params,
verbose=verbose,
ch=ch)
end
# For each value of the regularization parameter,
# compute the training error, ie, average error (sum over (i,j) in train_glrm.obs of L_j(A_ij, x_i y_j))
# and the test error, ie, average error (sum over (i,j) in test_glrm.obs of L_j(A_ij, x_i y_j))
function regularization_path(train_glrm::AbstractGLRM, test_glrm::AbstractGLRM; params=Params(), reg_params=exp10.(range(2,stop=-2,length=5)),
verbose=true,
ch::ConvergenceHistory=ConvergenceHistory("reg_path"))
train_error = Array{Float64}(undef, length(reg_params))
test_error = Array{Float64}(undef, length(reg_params))
ntrain = sum(map(length, train_glrm.observed_features))
ntest = sum(map(length, test_glrm.observed_features))
if verbose println("training model on $ntrain samples and testing on $ntest") end
@show params
train_time = Array{Float64}(undef, length(reg_params))
for iparam=1:length(reg_params)
reg_param = reg_params[iparam]
# evaluate train and test error
if verbose println("fitting train GLRM for reg_param $reg_param") end
scale_regularizer!(train_glrm, reg_param)
# no need to restart glrm X and Y even if they went to zero at the higher regularization
# b/c fit! does that automatically
fit!(train_glrm, params, ch=ch, verbose=verbose)
train_time[iparam] = ch.times[end]
if verbose println("computing mean train and test error for reg_param $reg_param:") end
train_error[iparam] = objective(train_glrm, parameter_estimate(train_glrm)..., include_regularization=false) / ntrain
if verbose println("\ttrain error: $(train_error[iparam])") end
test_error[iparam] = objective(test_glrm, parameter_estimate(train_glrm)..., include_regularization=false) / ntest
if verbose println("\ttest error: $(test_error[iparam])") end
end
return train_error, test_error, train_time, reg_params
end
function precision_at_k(train_glrm::GLRM, test_observed_features; params=Params(), reg_params=exp10.(range(2,stop=-2,length=5)),
holdout_proportion=.1, verbose=true,
ch::ConvergenceHistory=ConvergenceHistory("reg_path"), kprec=10)
m,n = size(train_glrm.A)
ntrain = sum(map(length, train_glrm.observed_features))
ntest = sum(map(length, test_observed_features))
train_observed_features = train_glrm.observed_features
train_error = Array{Float64}(undef, length(reg_params))
test_error = Array{Float64}(undef, length(reg_params))
prec_at_k = Array{Float64}(undef, length(reg_params))
solution = Array{Tuple{Float64,Float64}}(undef, length(reg_params))
train_time = Array{Float64}(undef, length(reg_params))
test_glrm = GLRM(train_glrm.A, train_glrm.losses, train_glrm.rx, train_glrm.ry, train_glrm.k,
X=copy(train_glrm.X), Y=copy(train_glrm.Y),
observed_features = test_observed_features)
for iparam=1:length(reg_params)
reg_param = reg_params[iparam]
# evaluate train error
if verbose println("fitting train GLRM for reg_param $reg_param") end
mul!(train_glrm.rx, reg_param)
mul!(train_glrm.ry, reg_param)
train_glrm.X, train_glrm.Y = randn(train_glrm.k,m), randn(train_glrm.k,n) # this bypasses the error checking in GLRM(). Risky.
X, Y, ch = fit!(train_glrm, params, ch=ch, verbose=verbose)
train_time[iparam] = ch.times[end]
if verbose println("computing train error and precision at k for reg_param $reg_param:") end
train_error[iparam] = objective(train_glrm, X, Y, include_regularization=false) / ntrain
if verbose println("\ttrain error: $(train_error[iparam])") end
test_error[iparam] = objective(test_glrm, X, Y, include_regularization=false) / ntrain
if verbose println("\ttest error: $(test_error[iparam])") end
# precision at k
XY = X'*Y
q = sort(XY[:],rev=true)[ntrain] # the ntest+ntrain largest value in the model XY
true_pos = 0; false_pos = 0
kfound = 0
for i=1:m
if kfound >= kprec
break
end
for j=1:n
if kfound >= kprec
break
end
if XY[i,j] >= q
# i predict 1 and (i,j) was in my test set and i observed 1
if j in test_observed_features[i]
true_pos += 1
kfound += 1
# i predict 1 and i did not observe a 1 (in either my test *or* train set)
elseif !(j in train_observed_features[i])
false_pos += 1
kfound += 1
end
end
end
end
prec_at_k[iparam] = true_pos / (true_pos + false_pos)
if verbose println("\tprec_at_k: $(prec_at_k[iparam])") end
solution[iparam] = (sum(X)+sum(Y), sum(abs.(X))+sum(abs.(Y)))
if verbose println("\tsum of solution, one norm of solution: $(solution[iparam])") end
end
return train_error, test_error, prec_at_k, train_time, reg_params, solution
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 2986 | # Supported domains: Real, Boolean, Ordinal, Periodic, Count
# The purpose of domains is to be able to impute over different possible values of `a` regardless of
# the loss that was used in the GLRM. The reason for doing this is to evaluate the performance of GLRMS.
# For instance, let's say we use PCA (QuadLoss losses) to model a binary data frame (not the best idea).
# In order to override the standard imputation with `impute(QuadLoss(), u)`, which assumes imputation over the reals,
# we can use `impute(BoolDomain(), QuadLoss(), u)` and see which of {-1,1} is best. The reason we want to be able to
# do this is to compare a baseline model (e.g. PCA) with a more logical model using heterogenous losses,
# yet still give each model the same amount of information regarding how imputation should be done.
# In order to accomplish this we define a series of domains that tell imputation methods
# what values the data can take. The imputation methods are defined in impute_and_err.jl
# Domains should be assigned to each column of the data and are not part of the low-rank model itself.
# They serve as a way to evaluate the performance of the low-rank model.
export Domain, # the abstract type
RealDomain, BoolDomain, OrdinalDomain, PeriodicDomain, CountDomain, CategoricalDomain, # the domains
copy
abstract type Domain end
########################################## REALS ##########################################
# Real data can take values from ℜ
struct RealDomain<:Domain
end
########################################## BOOLS ##########################################
# Boolean data should take values from {true, false}
struct BoolDomain<:Domain
end
########################################## ORDINALS ##########################################
# Ordinal data should take integer values ranging from `min` to `max`
struct OrdinalDomain<:Domain
min::Int
max::Int
function OrdinalDomain(min, max)
if max - min < 2
@warn("The ordinal variable you've created is degenerate: it has only two levels. Consider using a Boolean variable instead; ordinal loss functions may have unexpected behavior on a degenerate ordinal domain.")
end
return new(min, max)
end
end
########################################## ORDINALS ##########################################
# Categorical data should take integer values ranging from 1 to `max`
struct CategoricalDomain<:Domain
min::Int
max::Int
end
CategoricalDomain(m::Int) = CategoricalDomain(1,m)
########################################## PERIODIC ##########################################
# Periodic data can take values from ℜ, but given a period T, we should have error_metric(a,a+T) = 0
struct PeriodicDomain<:Domain
T::Float64 # the period
end
########################################## COUNTS ##########################################
# Count data can take values over ℕ, which we approximate as {0, 1, 2 ... `max_count`}
struct CountDomain<:Domain
max_count::Int # the biggest possible count
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 6254 | export objective, error_metric, impute, impute_missing
### OBJECTIVE FUNCTION EVALUATION FOR MPCA
function objective(glrm::GLRM, X::Array{Float64,2}, Y::Array{Float64,2},
XY::Array{Float64,2};
yidxs = get_yidxs(glrm.losses), # mapping from columns of A to columns of Y; by default, the identity
include_regularization=true)
m,n = size(glrm.A)
@assert(size(XY)==(m,yidxs[end][end]))
@assert(size(Y)==(glrm.k,yidxs[end][end]))
@assert(size(X)==(glrm.k,m))
err = 0.0
for j=1:n
for i in glrm.observed_examples[j]
err += evaluate(glrm.losses[j], XY[i,yidxs[j]], glrm.A[i,j])
end
end
# add regularization penalty
if include_regularization
err += calc_penalty(glrm,X,Y; yidxs = yidxs)
end
return err
end
function row_objective(glrm::AbstractGLRM, i::Int, x::AbstractArray, Y::Array{Float64,2} = glrm.Y;
yidxs = get_yidxs(glrm.losses), # mapping from columns of A to columns of Y; by default, the identity
include_regularization=true)
m,n = size(glrm.A)
err = 0.0
XY = x'*Y
for j in glrm.observed_features[i]
err += evaluate(glrm.losses[j], XY[1,yidxs[j]], glrm.A[i,j])
end
# add regularization penalty
if include_regularization
err += evaluate(glrm.rx[i], x)
end
return err
end
function col_objective(glrm::AbstractGLRM, j::Int, y::AbstractArray, X::Array{Float64,2} = glrm.X;
include_regularization=true)
m,n = size(glrm.A)
sz = size(y)
if length(sz) == 1 colind = 1 else colind = 1:sz[2] end
err = 0.0
XY = X'*y
obsex = glrm.observed_examples[j]
@inbounds XYj = XY[obsex,colind]
@inbounds Aj = convert(Array, glrm.A[obsex,j])
err += evaluate(glrm.losses[j], XYj, Aj)
# add regularization penalty
if include_regularization
err += evaluate(glrm.ry[j], y)
end
return err
end
# The user can also pass in X and Y and `objective` will compute XY for them
function objective(glrm::GLRM, X::Array{Float64,2}, Y::Array{Float64,2};
sparse=false, include_regularization=true,
yidxs = get_yidxs(glrm.losses), kwargs...)
@assert(size(Y)==(glrm.k,yidxs[end][end]))
@assert(size(X)==(glrm.k,size(glrm.A,1)))
XY = Array{Float64}(undef, (size(X,2), size(Y,2)))
if sparse
# Calculate X'*Y only at observed entries of A
m,n = size(glrm.A)
err = 0.0
for j=1:n
for i in glrm.observed_examples[j]
err += evaluate(glrm.losses[j], dot(X[:,i],Y[:,yidxs[j]]), glrm.A[i,j])
end
end
if include_regularization
err += calc_penalty(glrm,X,Y; yidxs = yidxs)
end
return err
else
# dense calculation variant (calculate XY up front)
gemm!('T','N',1.0,X,Y,0.0,XY)
return objective(glrm, X, Y, XY; include_regularization=include_regularization, yidxs = yidxs, kwargs...)
end
end
# Or just the GLRM and `objective` will use glrm.X and .Y
objective(glrm::GLRM; kwargs...) = objective(glrm, glrm.X, glrm.Y; kwargs...)
# For shared arrays
# TODO: compute objective in parallel
objective(glrm::ShareGLRM, X::SharedArray{Float64,2}, Y::SharedArray{Float64,2}) =
objective(glrm, X.s, Y.s)
# Helper function to calculate the regularization penalty for X and Y
function calc_penalty(glrm::AbstractGLRM, X::Array{Float64,2}, Y::Array{Float64,2};
yidxs = get_yidxs(glrm.losses))
m,n = size(glrm.A)
@assert(size(Y)==(glrm.k,yidxs[end][end]))
@assert(size(X)==(glrm.k,m))
penalty = 0.0
for i=1:m
penalty += evaluate(glrm.rx[i], view(X,:,i))
end
for f=1:n
penalty += evaluate(glrm.ry[f], view(Y,:,yidxs[f]))
end
return penalty
end
## ERROR METRIC EVALUATION (BASED ON DOMAINS OF THE DATA)
function raw_error_metric(glrm::AbstractGLRM, XY::Array{Float64,2}, domains::Array{Domain,1};
yidxs = get_yidxs(glrm.losses))
m,n = size(glrm.A)
err = 0.0
for j=1:n
for i in glrm.observed_examples[j]
err += error_metric(domains[j], glrm.losses[j], XY[i,yidxs[j]], glrm.A[i,j])
end
end
return err
end
function std_error_metric(glrm::AbstractGLRM, XY::Array{Float64,2}, domains::Array{Domain,1};
yidxs = get_yidxs(glrm.losses))
m,n = size(glrm.A)
err = 0.0
for j=1:n
column_mean = 0.0
column_err = 0.0
for i in glrm.observed_examples[j]
column_mean += glrm.A[i,j]^2
column_err += error_metric(domains[j], glrm.losses[j], XY[i,yidxs[j]], glrm.A[i,j])
end
column_mean = column_mean/length(glrm.observed_examples[j])
if column_mean != 0
column_err = column_err/column_mean
end
err += column_err
end
return err
end
function error_metric(glrm::AbstractGLRM, XY::Array{Float64,2}, domains::Array{Domain,1};
standardize=false,
yidxs = get_yidxs(glrm.losses))
m,n = size(glrm.A)
@assert(size(XY)==(m,yidxs[end][end]))
if standardize
return std_error_metric(glrm, XY, domains; yidxs = yidxs)
else
return raw_error_metric(glrm, XY, domains; yidxs = yidxs)
end
end
# The user can also pass in X and Y and `error_metric` will compute XY for them
function error_metric(glrm::AbstractGLRM, X::Array{Float64,2}, Y::Array{Float64,2}, domains::Array{Domain,1}=Domain[l.domain for l in glrm.losses]; kwargs...)
XY = Array{Float64}(undef,(size(X,2), size(Y,2)))
gemm!('T','N',1.0,X,Y,0.0,XY)
error_metric(glrm, XY, domains; kwargs...)
end
# Or just the GLRM and `error_metric` will use glrm.X and .Y
error_metric(glrm::AbstractGLRM, domains::Array{Domain,1}; kwargs...) = error_metric(glrm, glrm.X, glrm.Y, domains; kwargs...)
error_metric(glrm::AbstractGLRM; kwargs...) = error_metric(glrm, Domain[l.domain for l in glrm.losses]; kwargs...)
# Use impute and errors over GLRMS
impute(glrm::AbstractGLRM) = impute(glrm.losses, glrm.X'*glrm.Y)
function impute_missing(glrm::AbstractGLRM)
Ahat = impute(glrm)
for j in 1:size(glrm.A,2)
for i in glrm.observed_examples[j]
Ahat[i,j] = glrm.A[i,j]
end
end
return Ahat
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 1029 | export fit, fit!, Params
### PARAMETERS TYPE
abstract type AbstractParams end
Params(args...; kwargs...) = ProxGradParams(args...; kwargs...)
# default in-place fitting uses proximal gradient method
function fit!(glrm::AbstractGLRM; kwargs...)
kwdict = Dict(kwargs)
if :params in keys(kwdict)
return fit!(glrm, kwdict[:params]; kwargs...)
else
if isa(glrm.A,SparseMatrixCSC)
# Default to sparse algorithm for a sparse dataset
return fit!(glrm, SparseProxGradParams(); kwargs...)
else
# Classic proximal gradient method for non-sparse data
return fit!(glrm, ProxGradParams(); kwargs...)
end
end
end
# fit without modifying the glrm object
function fit(glrm::AbstractGLRM, args...; kwargs...)
X0 = Array{Float64}(undef, size(glrm.X))
Y0 = Array{Float64}(undef, size(glrm.Y))
copy!(X0, glrm.X); copy!(Y0, glrm.Y)
X,Y,ch = fit!(glrm, args...; kwargs...)
copy!(glrm.X, X0); copy!(glrm.Y, Y0)
return X',Y,ch
end
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |
|
[
"MIT"
] | 0.1.0 | 50e804aa999e452ce4bf5edc50493838bd744e09 | code | 8873 | # ========================================
# REVIEW THIS IN LIGHT OF NEW DATAFRAMES
# ========================================
import Base: isnan
import DataFrames: DataFrame, ncol, convert
export GLRM, observations, expand_categoricals!, NaNs_to_NAs!, NAs_to_0s!, NaNs_to_Missing!, ismissing_vec
include("fit_dataframe_w_type_imputation.jl")
probabilistic_losses = Dict{Symbol, Any}(
:real => QuadLoss,
:bool => LogisticLoss,
:ord => MultinomialOrdinalLoss,
:cat => MultinomialLoss
)
robust_losses = Dict{Symbol, Any}(
:real => HuberLoss,
:bool => LogisticLoss,
:ord => BvSLoss,
:cat => OvALoss
)
function GLRM(df::DataFrame, k::Int, datatypes::Array{Symbol,1};
loss_map = probabilistic_losses,
rx = QuadReg(.01), ry = QuadReg(.01),
offset = true, scale = false, prob_scale = true,
transform_data_to_numbers = true, NaNs_to_Missing = true)
# check input
if ncol(df)!=length(datatypes)
error("third argument (datatypes) must have one entry for each column of data frame.")
end
# validate input
for dt in datatypes
if !(dt in keys(loss_map))
error("data types must be either :real, :bool, :ord, or :cat, not $dt")
end
end
# clean up dataframe if needed
A = copy(df)
if NaNs_to_Missing
NaNs_to_Missing!(A)
end
# define loss functions for each column
losses = Array{Loss}(undef, ncol(A))
for j=1:ncol(df)
losstype = loss_map[datatypes[j]]
if transform_data_to_numbers
map_to_numbers!(A, j, datatypes[j])
end
losses[j] = pick_loss(losstype, A[:,j])
end
# identify which entries in data frame have been observed (ie are not missing)
obs = observations(df)
# form model
rys = Array{Regularizer}(undef, length(losses))
for i=1:length(losses)
if isa(losses[i].domain, OrdinalDomain) && embedding_dim(losses[i])>1 # losses[i], MultinomialOrdinalLoss) || isa(losses[i], OrdisticLoss)
rys[i] = OrdinalReg(copy(ry))
else
rys[i] = copy(ry)
end
end
glrm = GLRM(A, losses, rx, rys, k, obs=obs, offset=offset, scale=scale)
# scale model so it really computes the MAP estimator of the parameters
if prob_scale
prob_scale!(glrm)
end
return glrm
end
## transform data to numbers
function is_number_or_null(x)
isa(x, Number) || ismissing(x) # (:value in fieldnames(x) && isa(x.value, Number))
end
function is_int_or_null(x)
isa(x, Int) || ismissing(x) # (:value in fieldnames(x) && isa(x.value, Int))
end
function map_to_numbers!(df, j::Int, datatype::Symbol)
# easy case
if datatype == :real
if all(xi -> is_number_or_null(xi), df[:,j][.!ismissing_vec(df[:,j])])
return df[:,j]
else
error("column contains non-numerical values")
end
end
# harder cases
col = copy(df[:,j])
levels = Set(col[.!ismissing_vec(col)])
if datatype == :bool
if length(levels)>2
error("Boolean variable should have at most two levels; instead, got:\n$levels")
end
colmap = Dict{Any,Int}(zip(sort(collect(levels)), [-1,1][1:length(levels)]))
elseif datatype == :cat || datatype == :ord
colmap = Dict{Any,Int}(zip(sort(collect(levels)), 1:length(levels)))
else
error("datatype $datatype not recognized")
end
m = size(df,1)
df[!,j] = Array{Union{Missing, Int},1}(undef, m)
for i in 1:length(col)
if !ismissing(col[i])
df[i,j] = getval(colmap[col[i]])
end
end
return df[:,j]
end
getval(x::Union{T, Nothing}) where T = x.value
getval(x::T) where T<:Number = x
function map_to_numbers!(df, j::Int, loss::Type{QuadLoss})
if all(xi -> is_number_or_null(xi), df[:,j][!ismissing_vec(df[:,j])])
return df[:,j]
else
error("column contains non-numerical values")
end
end
function map_to_numbers!(df, j::Int, loss::Type{LogisticLoss})
col = copy(df[:,j])
levels = Set(col[!ismissing_vec(col)])
if length(levels)>2
error("Boolean variable should have at most two levels")
end
colmap = Dict{Any,Int}(zip(sort(collect(levels)), [-1,1][1:length(levels)]))
df[:,j] = DataArray(Int, length(df[:,j]))
for i in 1:length(col)
if !ismissing(col[i])
df[i,j] = colmap[col[i]]
end
end
return df[:,j]
end
function map_to_numbers!(df, j::Int, loss::Type{MultinomialLoss})
col = copy(df[:,j])
levels = Set(col[!ismissing_vec(col)])
colmap = Dict{Any,Int}(zip(sort(collect(levels)), 1:length(levels)))
df[:,j] = DataArray(Int, length(df[:,j]))
for i in 1:length(col)
if !ismissing(col[i])
df[i,j] = colmap[col[i]]
end
end
return df[:,j]
end
function map_to_numbers!(df, j::Int, loss::Type{MultinomialOrdinalLoss})
col = copy(df[:,j])
levels = Set(col[!ismissing_vec(col)])
colmap = Dict{Any,Int}(zip(sort(collect(levels)), 1:length(levels)))
df[:,j] = DataArray(Int, length(df[:,j]))
for i in 1:length(col)
if !ismissing(col[i])
df[i,j] = colmap[col[i]]
end
end
return df[:,j]
end
## sanity check the choice of loss
# this default definition could be tighter: only needs to be defined for arguments of types that subtype Loss
function pick_loss(l, col)
return l()
end
function pick_loss(l::Type{LogisticLoss}, col)
if all(xi -> ismissing(xi) || xi in [-1,1], col)
return l()
else
error("LogisticLoss can only be used on data taking values in {-1, 1}")
end
end
function pick_loss(l::Type{MultinomialLoss}, col)
if all(xi -> ismissing(xi) || (is_int_or_null(xi) && xi >= 1), col)
return l(maximum(skipmissing(col)))
else
error("MultinomialLoss can only be used on data taking positive integer values")
end
end
function pick_loss(l::Type{MultinomialOrdinalLoss}, col)
if all(xi -> ismissing(xi) || (isa(xi, Int) && xi >= 1), col)
return l(maximum(skipmissing(col)))
else
error("MultinomialOrdinalLoss can only be used on data taking positive integer values")
end
end
observations(da::Array{Union{T, Missing}}) where T = df_observations(da)
observations(df::DataFrame) = df_observations(df)
# isnan -> ismissing
function df_observations(da)
obs = Tuple{Int, Int}[]
m,n = size(da)
for j=1:n # follow column-major order. First element of index in innermost loop
for i=1:m
if !ismissing(da[i,j])
push!(obs,(i,j))
end
end
end
return obs
end
# TODO.. Missings in the data frame will be replaced by the number `z`
function df2array(df::DataFrame, z::Number)
A = zeros(size(df))
for i=1:size(A,2)
if issubtype(typeof(df[:,i]), Array)
A[:,i] = df[:,i]
elseif typeof(df[i]) == Bool
A[:,i] = convert(Array, (2*df[i]-1), z)
else
A[:,i] = convert(Array, df[i], z)
end
end
return A
end
df2array(df::DataFrame) = df2array(df, 0)
# expand categorical columns, given as column indices, into one boolean column for each level
function expand_categoricals!(df::DataFrame,categoricals::Array{Int,1})
# map from names to indices; not used: categoricalidxs = map(y->df.colindex[y], categoricals)
# create one boolean column for each level of categorical column
colnames = names(df)
for col in categoricals
levels = sort(unique(df[:,col]))
for level in levels
if !ismissing(level)
colname = Symbol(string(colnames[col])*"="*string(level))
df[colname] = (df[:,col] .== level)
end
end
end
# remove the original categorical columns
for cat in sort(categoricals, rev=true)
delete!(df, cat)
end
return df
end
function expand_categoricals!(df::DataFrame,categoricals::UnitRange{Int})
expand_categoricals!(df, Int[i for i in categoricals])
end
# expand categoricals given as names of columns rather than column indices
function expand_categoricals!(df::DataFrame,categoricals::Array)
# map from names to indices
categoricalidxs = map(y->df.colindex[y], categoricals)
return expand_categoricals!(df, categoricalidxs)
end
# convert NaNs to NAs
# isnan(x::NAtype) = false
isnan(x::AbstractString) = false
isnan(x::Union{T, Nothing}) where T = isnan(x.value)
# same functionality as above.
function NaNs_to_Missing!(df::DataFrame)
m,n = size(df)
for j=1:n
df[!,j] = [ismissing(df[i,j]) || isnan(df[i,j]) ? missing : value for (i,value) in enumerate(df[:,j])];
end
return df
end
ismissing_vec(V::AbstractArray) = Bool[ismissing(x) for x in V[:]]
| MatrixCompletion | https://github.com/jasonsun0310/MatrixCompletion.jl.git |