File size: 8,731 Bytes
618f7d3 88e7b46 618f7d3 b27bca9 cf880b5 b27bca9 cf880b5 618f7d3 cba6bb5 b27bca9 cba6bb5 b27bca9 618f7d3 cf880b5 b27bca9 cf880b5 b27bca9 cf880b5 b27bca9 618f7d3 dbce74b cba6bb5 dbce74b cf880b5 b27bca9 618f7d3 54c30e1 b27bca9 cf880b5 49f392e cf880b5 b27bca9 cf880b5 49f392e 618f7d3 67d834c cf880b5 49f392e cf880b5 49f392e 618f7d3 b27bca9 618f7d3 b27bca9 618f7d3 b27bca9 618f7d3 b27bca9 618f7d3 49f392e 618f7d3 cba6bb5 b27bca9 618f7d3 49f392e 618f7d3 b27bca9 618f7d3 49f392e 618f7d3 b27bca9 618f7d3 49f392e 618f7d3 54c30e1 cf880b5 67d834c 618f7d3 dbce74b 618f7d3 88e7b46 |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 |
---
tags:
- monai
- medical
library_name: monai
license: apache-2.0
---
# Model Overview
A neural architecture search algorithm for volumetric (3D) segmentation of the pancreas and pancreatic tumor from CT image. This model is trained using the neural network model from the neural architecture search algorithm, DiNTS [1].
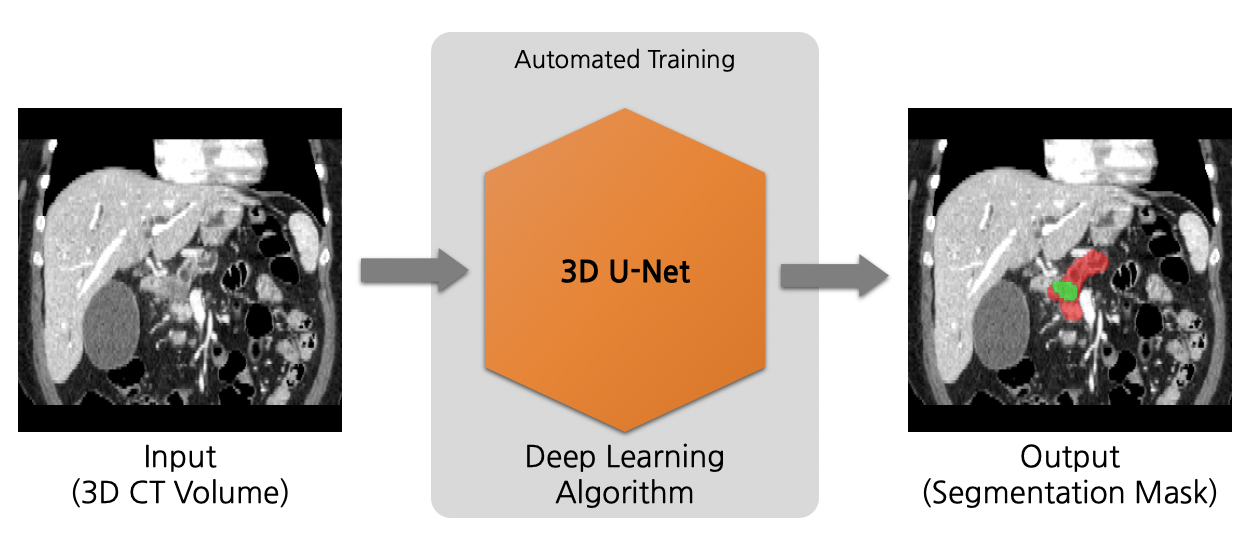
## Data
The training dataset is the Pancreas Task from the Medical Segmentation Decathalon. Users can find more details on the datasets at http://medicaldecathlon.com/.
- Target: Pancreas and pancreatic tumor
- Modality: Portal venous phase CT
- Size: 420 3D volumes (282 Training +139 Testing)
- Source: Memorial Sloan Kettering Cancer Center
- Challenge: Label unbalance with large (background), medium (pancreas) and small (tumour) structures.
### Preprocessing
The data list/split can be created with the script `scripts/prepare_datalist.py`.
```
python scripts/prepare_datalist.py --path /path-to-Task07_Pancreas/ --output configs/dataset_0.json
```
## Training configuration
The training was performed with at least 16GB-memory GPUs.
Actual Model Input: 96 x 96 x 96
### Neural Architecture Search Configuration
The neural architecture search was performed with the following:
- AMP: True
- Optimizer: SGD
- Initial Learning Rate: 0.025
- Loss: DiceCELoss
### Optimial Architecture Training Configuration
The training was performed with the following:
- AMP: True
- Optimizer: SGD
- (Initial) Learning Rate: 0.025
- Loss: DiceCELoss
The segmentation of pancreas region is formulated as the voxel-wise 3-class classification. Each voxel is predicted as either foreground (pancreas body, tumour) or background. And the model is optimized with gradient descent method minimizing soft dice loss and cross-entropy loss between the predicted mask and ground truth segmentation.
### Input
One channel
- CT image
### Output
Three channels
- Label 2: pancreatic tumor
- Label 1: pancreas
- Label 0: everything else
### Memory Consumption
- Dataset Manager: CacheDataset
- Data Size: 420 3D Volumes
- Cache Rate: 1.0
- Multi GPU (8 GPUs) - System RAM Usage: 400G
### Memory Consumption Warning
If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate `cache_rate` in the configurations within range [0, 1] to minimize the System RAM requirements.
## Performance
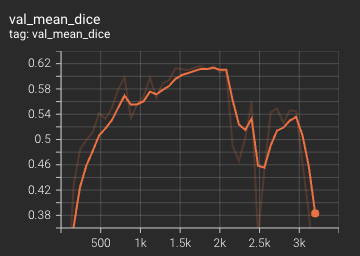
Dice score is used for evaluating the performance of the model. This model achieves a mean dice score of 0.62.
Please note that this bundle is non-deterministic because of the trilinear interpolation used in the network. Therefore, reproducing the training process may not get exactly the same performance.
Please refer to https://pytorch.org/docs/stable/notes/randomness.html#reproducibility for more details about reproducibility.
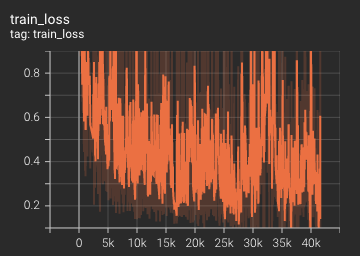
#### Training Loss
The loss over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
#### Validation Dice
The mean dice score over 3200 epochs (the bright curve is smoothed, and the dark one is the actual curve)
#### TensorRT speedup
This bundle supports acceleration with TensorRT. The table below displays the speedup ratios observed on an A100 80G GPU.
| method | torch_fp32(ms) | torch_amp(ms) | trt_fp32(ms) | trt_fp16(ms) | speedup amp | speedup fp32 | speedup fp16 | amp vs fp16|
| :---: | :---: | :---: | :---: | :---: | :---: | :---: | :---: | :---: |
| model computation | 54611.72 | 19240.66 | 16104.8 | 11443.57 | 2.84 | 3.39 | 4.77 | 1.68 |
| end2end | 133.93 | 43.41 | 35.65 | 26.63 | 3.09 | 3.76 | 5.03 | 1.63 |
Where:
- `model computation` means the speedup ratio of model's inference with a random input without preprocessing and postprocessing
- `end2end` means run the bundle end-to-end with the TensorRT based model.
- `torch_fp32` and `torch_amp` are for the PyTorch models with or without `amp` mode.
- `trt_fp32` and `trt_fp16` are for the TensorRT based models converted in corresponding precision.
- `speedup amp`, `speedup fp32` and `speedup fp16` are the speedup ratios of corresponding models versus the PyTorch float32 model
- `amp vs fp16` is the speedup ratio between the PyTorch amp model and the TensorRT float16 based model.
This result is benchmarked under:
- TensorRT: 8.6.1+cuda12.0
- Torch-TensorRT Version: 1.4.0
- CPU Architecture: x86-64
- OS: ubuntu 20.04
- Python version:3.8.10
- CUDA version: 12.1
- GPU models and configuration: A100 80G
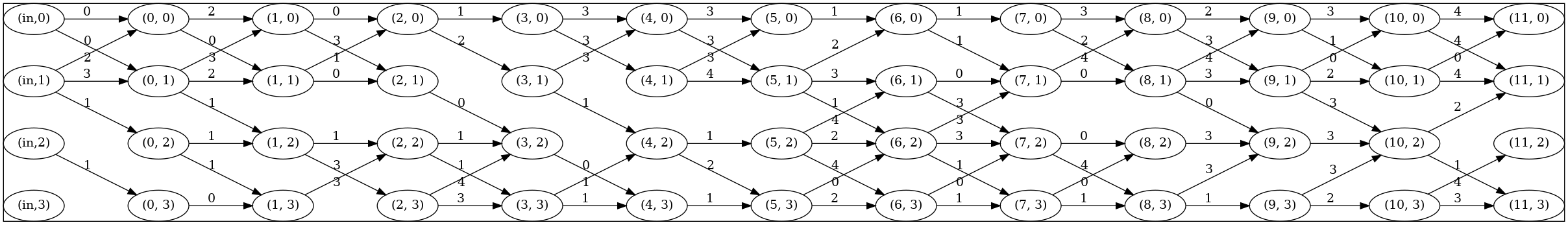
### Searched Architecture Visualization
Users can install Graphviz for visualization of searched architectures (needed in [decode_plot.py](https://github.com/Project-MONAI/tutorials/blob/main/automl/DiNTS/decode_plot.py)). The edges between nodes indicate global structure, and numbers next to edges represent different operations in the cell searching space. An example of searched architecture is shown as follows:
## MONAI Bundle Commands
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file.
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html).
#### Execute model searching:
```
python -m scripts.search run --config_file configs/search.yaml
```
#### Execute multi-GPU model searching (recommended):
```
torchrun --nnodes=1 --nproc_per_node=8 -m scripts.search run --config_file configs/search.yaml
```
#### Execute training:
```
python -m monai.bundle run --config_file configs/train.yaml
```
Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using `--dataset_dir`:
```
python -m monai.bundle run --config_file configs/train.yaml --dataset_dir <actual dataset path>
```
#### Override the `train` config to execute multi-GPU training:
```
torchrun --nnodes=1 --nproc_per_node=8 -m monai.bundle run --config_file "['configs/train.yaml','configs/multi_gpu_train.yaml']"
```
#### Override the `train` config to execute evaluation with the trained model:
```
python -m monai.bundle run --config_file "['configs/train.yaml','configs/evaluate.yaml']"
```
#### Execute inference:
```
python -m monai.bundle run --config_file configs/inference.yaml
```
#### Export checkpoint for TorchScript:
```
python -m monai.bundle ckpt_export network_def --filepath models/model.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.yaml
```
#### Export checkpoint to TensorRT based models with fp32 or fp16 precision:
```
python -m monai.bundle trt_export --net_id network_def --filepath models/model_trt.ts --ckpt_file models/model.pt --meta_file configs/metadata.json --config_file configs/inference.yaml --precision <fp32/fp16> --use_trace "True" --dynamic_batchsize "[1, 4, 8]" --converter_kwargs "{'truncate_long_and_double':True, 'torch_executed_ops': ['aten::upsample_trilinear3d']}"
```
#### Execute inference with the TensorRT model:
```
python -m monai.bundle run --config_file "['configs/inference.yaml', 'configs/inference_trt.yaml']"
```
# References
[1] He, Y., Yang, D., Roth, H., Zhao, C. and Xu, D., 2021. Dints: Differentiable neural network topology search for 3d medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (pp. 5841-5850).
# License
Copyright (c) MONAI Consortium
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
|